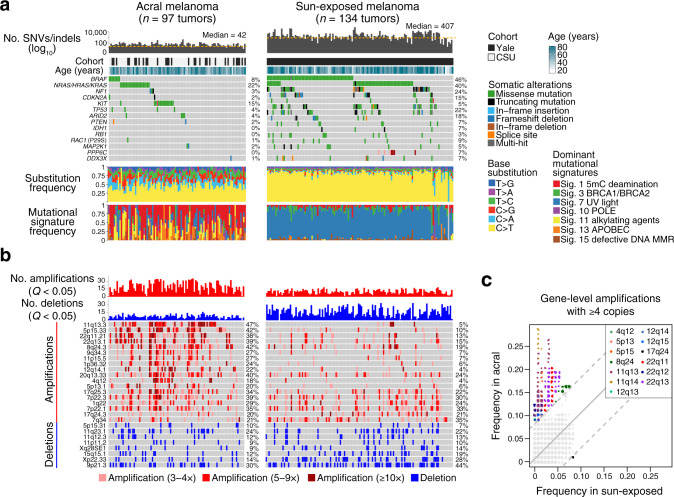
Fig. 1. Landscape of somatic alterations in acral and sun-exposed melanomas.
a, b Genomic and clinical characterization of acral and sun-exposed melanoma samples sequenced in this work. a The number of nonsynonymous SNVs and indels per melanoma exome (columns), cohort, age, frequently mutated genes in either subtype (Methods), nonsynonymous base substitution frequencies, and dominant COSMIC mutational signatures122, 123. Sig. signature, 5mC 5-methylcytosine. See also Supplementary Data 2b, c for the results of mutational significance analysis with MutSigCV102. b The number of significant focal amplifications and deletions (GISTIC Q < 0.05) per melanoma exome (columns), ordered identically to panel a. Cytobands with focal amplifications or deletions with at least 10% recurrence frequency in either melanoma subtype are shown (GISTIC Q < 10–5), ordered by the relative difference in recurrence frequency in acral versus sun-exposed melanoma. c Genes are plotted according to the fraction of acral (y-axis) or sun-exposed (x-axis) tumors where they are present with ≥4 copies. Considering the genome-wide distribution of differences in recurrence frequencies between melanoma subtypes, genes are identified as significantly recurrent in acral or sun-exposed melanomas if their |z-score | ≥ 3 (dashed lines). Significantly recurrent genes are colored according to their cytoband location (inset). For clarity, a small amount of jitter was added to distinguish overlapping genes. Source data are provided as a Source Data file.