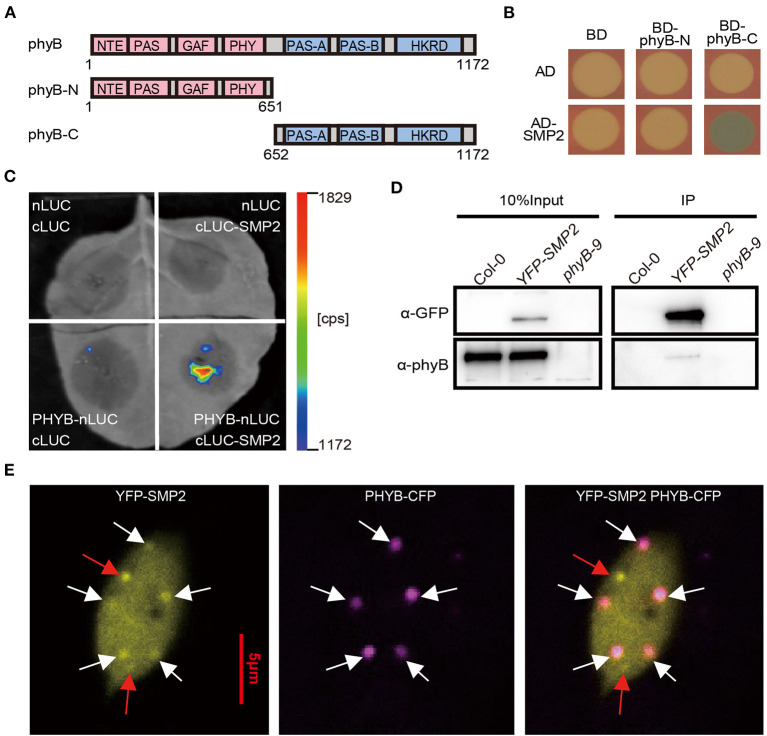
Figure 1.
phyB physically interacts with SMP2 in vitro and in vivo. (A) Schematic diagram of phyB fragments. Numbers indicate the amino acid positions in phyB protein. (B) The interaction of SMP2 and phyB in yeast. AD, activation domain; BD, DNA-binding domain. (C) Firefly luciferase complementation imaging (LCI) assay showing interaction between phyB and SMP2 in tobacco leaf. nLUC, the N-terminal fragment of firefly luciferase (LUC); cLUC, the C-terminal fragment of LUC. Full-length phyB and SMP2 were fused to the nLUC and cLUC, respectively. (D) Co-IP assay showing the association between SMP2 and phyB. Seedlings grown in the dark were transferred to red light (145 μmol/m2·s) for 1 h. YFP-SMP2 proteins were pulled down with GFP-trap beads. α-GFP, anti-GFP antibody; α-phyB, anti-phyB antibody. (E) Colocalization analysis of SMP2 and phyB in vivo. Transgenic plants co-expressing YFP-SMP2 and phyB-CFP were grown in continuous red light (145 μmol/m2·s) for 5 days. The images of nucleus came from a hypocotyl cell. YFP-SMP2 fusion proteins were excited by laser at 514 nm, and the emitted fluorescence signaling was collected from 519 nm to 620 nm; phyB-CFP were excited by laser at 405 nm, and the emitted fluorescence was collected from 410 nm to 513 nm. Scale bar, 5 μm. White arrowheads indicate SMP2 and phyB colocalized in photobodies; Red arrowheads indicate the SMP2-specific nuclear speckles.