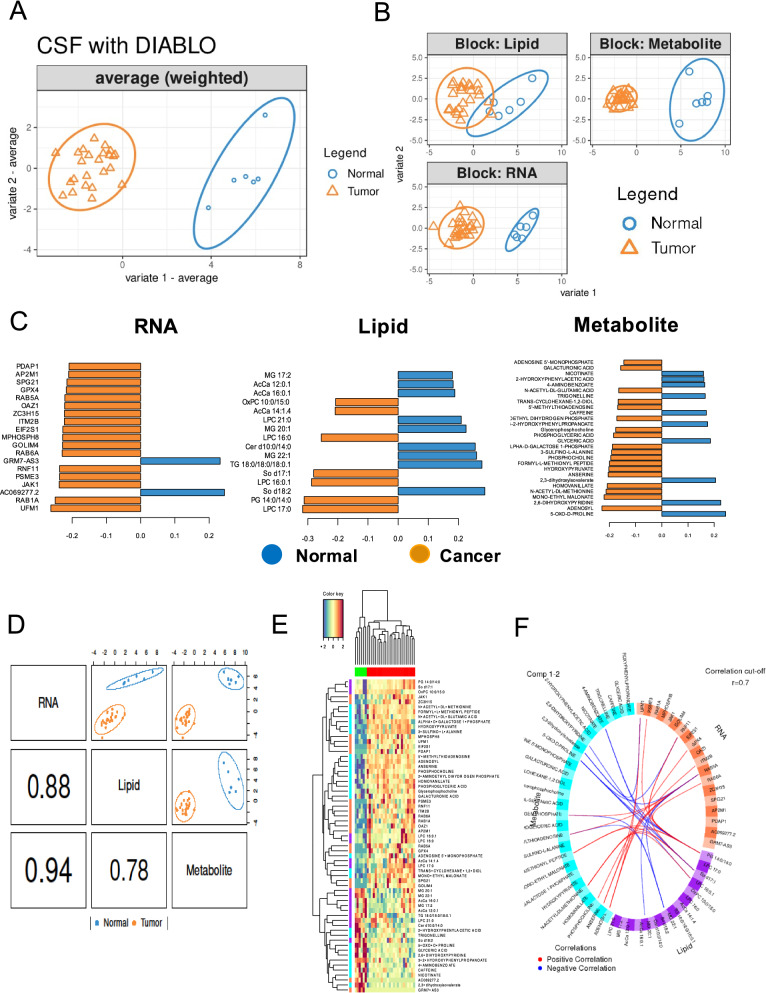
Fig. 5.
Sparse partial least-squares discriminant analysis. A sPLS-DA consensus plot for the combination of the three datasets showing complete discrimination of the 30 CSF samples (24 medulloblastoma and six normal samples). B The individual contribution of each dataset to the sPLS-DA final model, in each case showing the score plots for the two first components, indicating the best separation capability for transcriptome data followed by metabolome and lipidome data. C Selected features shown in pyramid bar plot. Loading plot represents the top 19 RNAs, 28 metabolites, and 16 lipids contributing to group separation. D Sample scatterplot from plotDiablo displaying the first component in each dataset (upper diagonal plot) and Pearson correlation between each component (lower diagonal plot). E Clustered image map (Euclidean distance, complete linkage) of the multi-omics signature based on the 54 multi-omics signature identified on the first component. Samples are represented in rows, selected features on the first component in columns. F The Circos plot (cut off: 0.7) shows positive or negative correlations denoted as red and blue lines, respectively