Abstract
The mortality rates among patients who initially survive sepsis are, in part, associated with a high risk of secondary lung infections and respiratory failure. Given that phagolysosomes are important for intracellular killing of pathogenic microbes, we investigated how severe lung infections associated with post-sepsis immunosuppression affect phagolysosome biogenesis. In mice with P. aeruginosa-induced pneumonia, we found a depletion of both phagosomes and lysosomes, as evidenced by decreased amounts of microtubule associated protein light chain 3-II (LC3-II) and lysosomal-associated membrane protein (LAMP1). We also found a loss of transcription factor E3 (TFE3) and transcription factor EB (TFEB), which are important activators for transcription of genes encoding autophagy and lysosomal proteins. These events were associated with increased expression of ZKSCAN3, a repressor for transcription of genes encoding autophagy and lysosomal proteins. Zkscan3−/− mice had increased expression of genes involved in the autophagy-lysosomal pathway along with enhanced killing of P. aeruginosa in the lungs, as compared to wild-type mice. These findings highlight the involvement of ZKSCAN3 in response to severe lung infection, including susceptibility to secondary bacterial infections due to immunosuppression.
Keywords: autophagy, transcription factors, bacteria, lung injury
Introduction
Despite advances in critical care, infections and trauma-related sepsis are leading causes of in-hospital mortality 1,2. While supportive interventions such as antibiotics, oxygen, and fluid resuscitation are frequently applied, effective therapeutics are not available for more than 200,000 critically-ill patients. Previous clinical trials, including those targeting coagulation and selected inflammatory mediators, appeared to have modest or no effect for sepsis survival. It is important to note that mortality rates are even greater among patients that initially survive sepsis but developed post-sepsis complications 3. In particular, onset of immunosuppression is related to a high risk of pneumonia and respiratory dysfunction. Therapeutic interventions for post-sepsis immunosuppression are not available for more than two and half million sepsis survivors in the US 4–7. Understanding the mechanisms involved in immune and lung tissue responses to sepsis and life-threatening secondary infections are crucial for development of effective interventions for sepsis survivors.
The development of new therapeutic strategies are essential to prevent reoccurrence of infection and organ injury by opportunistic pathogens during immunosuppression. Although a significant body of research has been performed connecting sepsis, and inflammation to redox biology, including the role of nitric oxide and hydrogen sulfide the integrated regulation of these pathways remains poorly defined 8–10. We and others have proposed that autophagy is an essential antioxidant pathway and since proteotoxicity and oxidative stress are central features of sepsis, we and others have proposed that autophagy also plays a critical role 11. In addition, given that intracellular killing of pathogenic microbes is largely mediated by phagolysosomes, preventing phagolysosome inactivation may be an important strategy to attenuate lung infections and immunosuppression 12–15. Phagolysosomes are a result of fusion between phagosomes and lysosomes that allows for effective degradation of many regulatory and damaged proteins, as well as neutralization of dysfunctional organelles, in processes such as mitochondrial quality control 16–18. Importantly, this degradative mechanism is also utilized for pathogen neutralization 12,19. Phagolysosome formation is a complex process that is initiated by LC3-I lipidation into LC3-II followed by incorporation into the phagosomal membranes 16. In addition to post-translational modifications, phagolysosomes are regulated at the transcriptional level 20–24. In particular, TFEB is known to promote expression of many autophagy and lysosomal genes, including LC3 and LAMP1 20,25. This is possible because TFEB recognizes the Coordinated Lysosomal Expression and Regulation (CLEAR) consensus sequence that is shared among many genes that encode autophagy and lysosomal components 26. In contrast to TFEB, transcriptional repression is mediated by zinc finger containing KRAB and SCAN domains 3 (ZKSCAN3) protein 27,28. ZKSCAN3 inhibits transcription of lysosomal genes and thereby effectively diminishing lysosome biogenesis 29–31. Recent studies indicate the capacity of ZKSCAN3 to block expression of autophagy-lysosome mediators in cancer cells 32–35. Although altered function of TFEB and also TFE3 have been demonstrated in inflammatory conditions 36, their role in regulating phagosome, and lysosome biogenesis during severe infections remains to be determined. Given that the role of ZKSCAN3 in bacterial infection has not been investigated, we hypothesized that ZKSCAN3 accumulation versus its deficiency will affect the severe bacterial infections and post-sepsis complications related to immunosuppression. Here we provide novel evidence of increased ZKSCAN3 levels in response to both bacterial infection and post-septic immunosuppression, and that ZKSCAN3 deficient mice (Zkscan3−/−) have enhanced bacterial killing during P. aeruginosa-induced pneumonia.
Materials and Methods
Mice.
Male C57BL/6 mice, 10–12 weeks old, were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were kept on a 12-hour light/dark cycle with free access to food and water. All experiments were conducted in accordance with approved protocols by the University of Alabama at Birmingham, Institutional Animal Care and Use Committee (IACUC). ZKSCAN3 knockout mice were generated by the Ingenious Targeting laboratory. First, a 12.2 Kb region of the Zkscan3 gene (RP23–346F12 C57BL/6 BAC clone) was used to construct the targeting vector. The short homology arm exten ds ~3.17 Kb 5’ to the Neo cassette (PGK-gb2-Neo). The long homology arm ends 3’ to the single LoxP site and is 5.8 Kb long. The loxP/FRT flanked Neo cassette was inserted 156 bp upstream of exon 3. The single LoxP site is inserted 2.14 Kb downstream of exon 5. The target region is 3.25 Kb and includes exons 3–5. Targeted iTK HF4 (129/SvEv x C57BL/6 FLP) hybrid embryonic stem cells were microinjected into C57BL/6 blastocysts. Germline Neo Deletion and FLP absence was confirmed to generate Zkscan3f/f mice. After breeding Zkscan3f/f with B6N.FVB-Tmem163Tg(ACTB-cre)2Mrt/CjDswJ (β-actin-cre) (Jackson Laboratory), the exons 3–5 were deleted. Mice were bred to C57/BL6 background for >8 generations. Primers for genotyping are: NDEL1: AGG CCA TGC CTT AAT GGG TGG, NDEL2: TGA TGT CAA CAG CAC TGC CTT GG; and SC1: GGT TTG GTT TTG CCT GGT GCA AAT G.
Reagents and antibodies.
LPS derived from E. coli 0111:B4 (L2630) and RIPA buffer (R0278) were purchased from Sigma (St. Louis, MO), Luria-Bertani broth (L8050) Luria-Bertani Agar+50μg/ml Ampicillin (L1002) were from Teknova (Hollister, CA). PBS (10010–023) was obtained from Thermo Fisher Scientific (Waltham, MA), whereas saline (MSDW-1000) was from Growcells (Irvine, CA). Western blot analysis was conducted using specific antibodies to LAMP1 (Ab25245; Abcam Cambridge, MA), TFEB (A303–673A; Bethyl, Montgomery, TX), and ZKSCAN3 (SC515285; Santa Cruz, Dallas TX). Antibodies to TFE3 (HPA023881), LC3 (L8918) and β-actin (A5441) were from Sigma (St. Louis, MO). Antibodies for immunohistochemistry are as the follows: Rubicon: Proteintech 21444–1-AP.
Mouse model of bacterial pneumonia.
P. aeruginosa strain K (PAK) was prepared in PBS (109 bacteria/ml), as described previously 37. Mice were anesthetized with isoflurane inhalation and suspended by their upper incisors on a 60o incline board. The tongue was gently extended followed by oropharyngeal deposition and aspiration of PBS (50 μl, control) or PBS containing P. aeruginosa strain K (PAK; 2.5 × 107/ml). The selected bacterial dose resulted in 100% mortality in 16 hours; therefore, lung homogenates were prepared 4 hours after P. aeruginosa instillation and serial dilutions used to determine colony-forming units (CFUs) on agar plates 38.
Mouse model of LPS-induced acute lung injury.
Our previous studies have shown that endotoxin (LPS) induced acute lung injury is characterized by neutrophil lung infiltration, interstitial edema, and accumulation of pro-inflammatory cytokines 39. Similar to our previous studies, mice (control) were treated with of saline (50 μl) or received saline and with LPS (2 mg/kg; i.t.). Mice were euthanized 24 h after LPS administration and lung homogenates prepared for western blot analysis 39.
Mouse model of sepsis-induced immunosuppression.
Cecal Ligation and Puncture (CLP) was performed as previously described 40,41. In brief, the cecum was ligated 25% from the tip, which results in an approximately 25% of the cecum ligated. A through-and-through puncture was performed with a 21-gauge needle and then a drop of feces was extruded to the peritoneal cavity. Saline (0.9%; 500 μl) was then applied into the peritoneal cavity and the abdominal wall incision was closed in two layers. The control group of mice (sham) underwent surgery without CLP. Post-CLP mice developed immunosuppression, including increase severity of lung infections confirmed by intratracheal instillation of P. aeruginosa, as previously described in our recent studies 19. Mice were euthanized 7 days after surgery and lung homogenates stored in liquid nitrogen for further analyses.
Cytokine ELISA:
Bronchoalveolar lavage (BAL) fluids were obtained by cannulating the trachea with a blunt 20-gauge needle and then lavaging the lungs three times with 1 ml of cold PBS. The amount of TNF-α and MIP-were determined using commercially available ELISA kits (R&D Systems) according to the manufacturer’s instructions.
Western blot analysis.
Cells or tissues were prepared in RIPA buffer supplemented with protease/phosphatase inhibitor cocktail (Thermofisher cat #1861284). The DC Lowry (Bio-Rad) assay was used to determine protein quantification and 10–20 μg protein per lane were loaded onto 10 or 12% PAGE SDS gels. After separation, proteins were transferred to PVDF or nitrocellulose membrane to probe for antibodies indicated in Reagents and antibodies section. Each experiment was carried out from lung homogenates obtained from indicated groups of mice (n = 3–5).
Quantitative real-time PCR analyses.
RNA was prepared from the lung tissue using Trizol followed by cDNA synthesis using High-Capacity cDNA Reverse Transcription Kit (Thermal Fisher Scientific, (Waltham, MA). Quantitative real-time PCR was performed with SYBR Green Mastermix (Thermal Fisher Scientific, Waltham, MA) with the following conditions: 95ºC; 5 min.; and then 40 cycles at 95ºC; 10 sec.; 60ºC, 10 sec.; 72ºC; 15 sec. Results were normalized to the levels of β-actin. Forward (F) and reverse (R) primer sequences are depicted in Table 1:
Table 1.
Primer sets for indicated gene expression.
| Forward | Reverse | |
|---|---|---|
| Atg5 | TCTGCAGGACAGTGTGATCC | GGCTAAGTTCTTCCTTCAACCA |
| Becn1 | AGCCTCTGAAACTGGACACG | CTTCCTCCTGGGTCTCTCCT |
| Cat | GCTCCGCAATCCTACACCAT | GGTGGTCAGGACATCAGGTC |
| Ctsd | CCGGTCTTTGACAACCTGAT | TCAGTGCCACCAAGCATTAG |
| Drp1 | TGCCTCAGATCGTCGTAGTG | CACCAGTTCCTCTGGGAAGA |
| G6pc | GATGCCTTCCACCAAGCTGA | GTAGAAGGCCATCCCGGAAC |
| Gclc | ACAAGGACGTGCTCAAGTGG | GTCTCAAGAACATCGCCTCCA |
| Gclm | GGAACCTGCTCAACTGGGG | GGGCTGATTTGGGAACTCCA |
| Gsr | CGGTGGTGGAGAGTCACAAG | TGAATTCCGAGTGCACTGCT |
| Lamp1 | CTGTCGAGTGGCAACTTCAG | GGATACAGTGGGGTTTGTGG |
| Lamp2a | TGGCTAATGGCTCAGCTTTC | ATGGGCACAAGGAAGTTGTC |
| Map1-lc3 | GTGGAAGATGTCCGGCTCAT | TGGTCAGGCACCAGGAACTT |
| Mcoln1 | GCGCGCCTCAGAGACTGA | TCTTCCTCTTCTGTGGGGGT |
| Nqo1 | CTCCCTCAACATCTGGAGCC | GCCTCCTTCATGGCGTAGTT |
| Sqstm1/P62 | CGAGTGGCTGTGCTGTTC | TGTCAGCTCCTCATCACTGG |
| Prkn | GAGCTTCCGAATCACCTGAC | CCCTCCAGATGCATTTGTTT |
| Pgc1-α | ACAGCTTTCTGGGTGGATTG | TCTGTGAGAACCGCTAGCAA |
| Rubcn | GATGGGGAGCGTCTGCTA | GTTGGCTGACACCAAACCTT |
| Sod1 | GGAAGCATGGCGATGAAAGC | CACAACTGGTTCACCGCTTG |
| Sod2 | GGCCAAGGGAGATGTTACAA | GCTTGATAGCCTCCAGCAAC |
| Tfe3 | CATCCTCGATCCTGACAGCT | TGTGGCCTGCAGTGATATTGG |
| Tfeb | GCGGACAGATTGACCTTCAG | CTCTCGCTGCTCCTCCTG |
| Wipi2 | GCTGTTCGCCAACTTCAAC | TCTGTTCCAGCTTATCCACAGA |
| Zkscan3 | CTGGATGACACACCTCCACA | TTTCTGAGCCCTGAGCTGAT |
| Actb | GACGGCCAGGTCATCACTAT | AAGGAAGGCTGGAAAAGAGC |
H&E and Immunohistochemistry:
Mouse lungs were fixed by inflation and submersion for 24 hrs in 4% paraformaldehyde/ 1% glutaraldehyde solution and embedded in paraffin. Sections (5 μm) were either stained with H&E or deparaffinized in serial solutions of Citrisolv (Fisher Scientific), isopropyl alcohol and water, followed by antigen retrieval by steaming in 10 mM citric acid (pH 6.0) for 20 minutes and then by a 20 minutes cooling period. After blocking endogenous peroxidase by placing slides in 3% hydrogen peroxide for 10 minutes, sections were washed with PBS buffer and blocked with 3% BSA for 60 min, and incubated with anti-Rubicon or anti-ATG5 antibody overnight at 4°C and then biotin-conjugated secondary antibody for 30 min. After staining with DAB, nuclei were counterstained with Hematoxylin. Sections were dehydrated and mounted with Permount. Images were taken using Keyence BZ-x810 microscope.
Statistical analysis.
Statistical analysis was performed using three or more independent experiments. Multigroup comparisons were obtained using one-way ANOVA with Tukey’s post hoc test. Values were normally distributed. Statistical significance was determined by the Student’s t-test for comparisons between two groups. A value of p < 0.05 was considered significant. Analyses were performed on GraphPad Prism 8.2.1. (GraphPad software, Inc, La Jolla, CA).
Results
Severe infections led to depletion of phagolysosomes and lysosomes in lungs of mice.
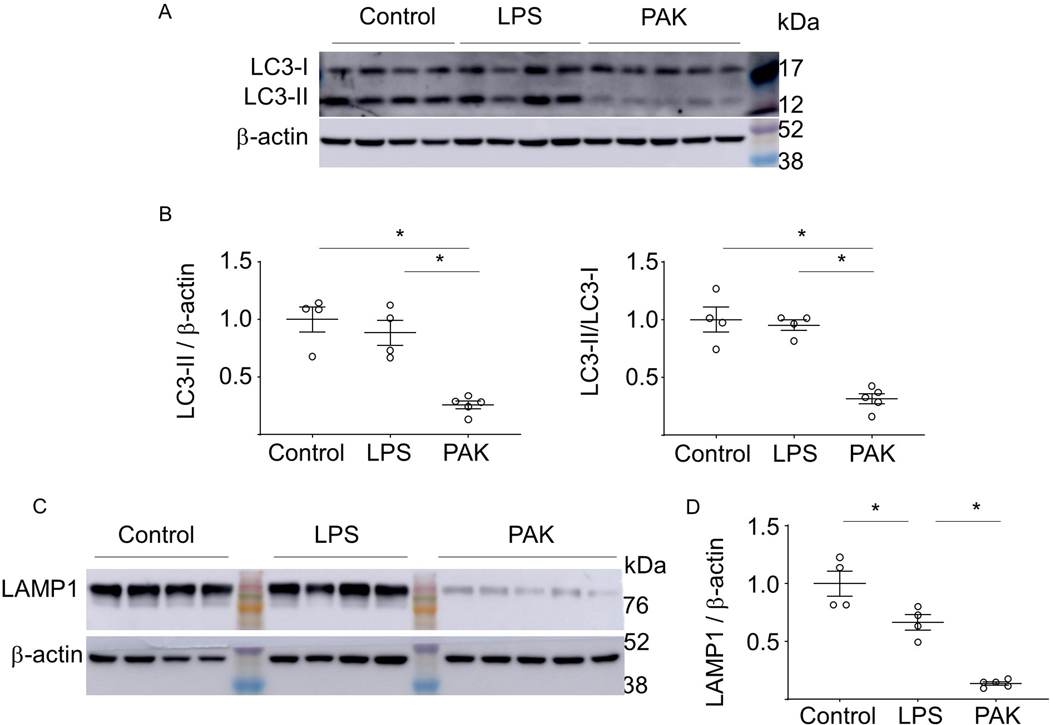
We tested whether sterile inflammatory conditions or lethal bacterial infections affect phagolysosomes and lysosomes in murine lungs. The LC3-II level as well as the LC3-II/LC3-I ratio were determined in control (unstimulated) or lungs from mice treated with LPS (2 mg/kg; i.t.) for 24 hours or P. aeruginosa strain K (PAK, 2.5 × 107/mouse; i.t.) for 4 hours. Western blot analysis indicated that the LC3-II and the LC3-II/LC3-I ratio were significantly decreased after exposure to PAK, while LPS had little or no effect (Fig. 1A and B). Similarly, PAK but not LPS decreased the amounts of LAMP1 that is an important component of the lysosomal membrane (Fig. 1C and D). The decrease of both LC3-II and LAMP1 indicate that severe lung infections lead to phagolysosome and lysosome depletion, while sterile inflammatory conditions induced by LPS had only a modest impact.
Figure 1.
The effects of endotoxin-induced acute lung injury and P. aeruginosa-mediated pneumonia on autophagy, phagolysosomal and lysosomal regulatory components in mouse lung. (A) Representative Western blots show LC3-I, LC3-II and β-actin levels in lung homogenates from control and mice subjected to intratracheal (i.t.) instillation of LPS for 24 hours, or after exposure to P. aeruginosa strain K (PAK) for 4 hours. The molecular weight markers (kDa) are indicated. (B) Quantitative analysis of LC3-II/β-actin and LC3-II/LC3-I ratios. Data = mean ± sem, n = 4–5, *p < 0.05 (ANOVA). (C) Western blots and (D) quantitative analysis of LAMP1 in lung homogenates obtained from indicated groups of mice. Data = mean ± sem, n = 4–5, *p < 0.05 (ANOVA).
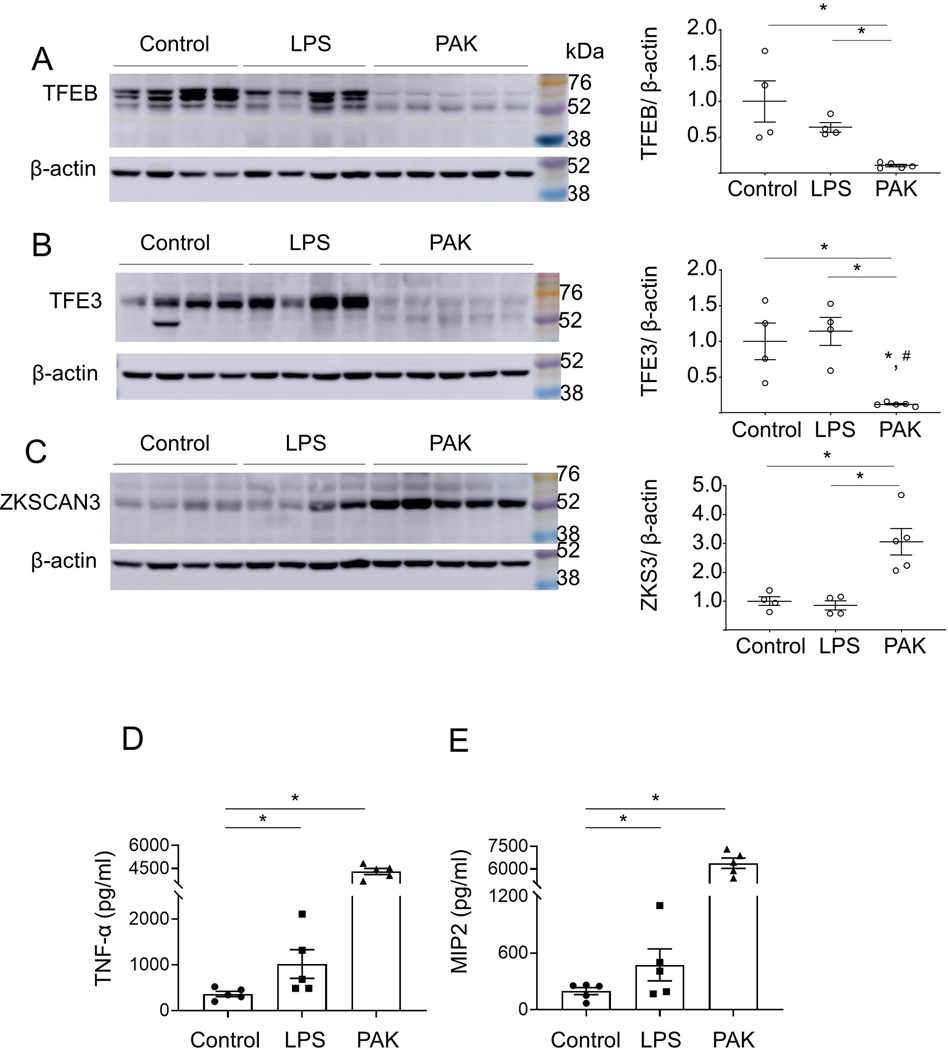
Increased ZKSCAN3 and decreased TFEB/TFE3 in lungs of mice with P. aeruginosa -induced pneumonia.
Recent studies have shown that crosstalk between activators TFEB /TFE3 and inhibitor ZKSCAN3 can affect phagolysosome and lysosome biogenesis in cancer cells 26,32,42. We found that both TFEB and TFE3 are diminished in lungs of PAK-infected mice, as compared to control group or mice subjected to LPS (i.t.) (Fig. 2A and B). Notably, the suppression of TFEB and TFE3 levels was accompanied by a robust increase of the transcriptional repressor ZKSCAN3, as evidenced by western blot analysis of these transcriptional regulators in lung homogenates (Fig. 2A–C). The amounts of TNF-α and MIP2 were significantly increased in the bronchoalveolar lavages (BALs) for either LPS or PAK compared to control groups of mice. Notably, a robust ~3-fold higher accumulation of these cytokines was observed after exposure to PAK for 4 hours, as compared to LPS-treated mice for 24 hours (Fig. 2D and E). These results indicate that severe lung infections are associated with accumulation of ZKSCAN3, while TFEB and TFE3 were decreased.
Figure 2.
The effects of endotoxin-induced acute lung injury and P. aeruginosa–induced pneumonia on TFEB, TFE3 and ZKSCAN3 levels in lungs of mice. Representative Western blots and optical densitometry show (A) TFEB, (B) TFE3, and (C) ZKSCAN3 protein levels in lung homogenates from indicated groups of mice. Samples were prepared 24 hours post-LPS (2 mg/kg; i.t.) or 4 hours after exposure to PAK (2.5 × 107; i.t.). Data = mean ± sem, n = 4–5, *p < 0.05 (ANOVA). (D) The amounts of TNF-α and (E) MIP2 cytokines in bronchoalveolar lavages (BALs) of indicated groups of mice. Mean ± sem, n = 5, *p < 0.05 (ANOVA).
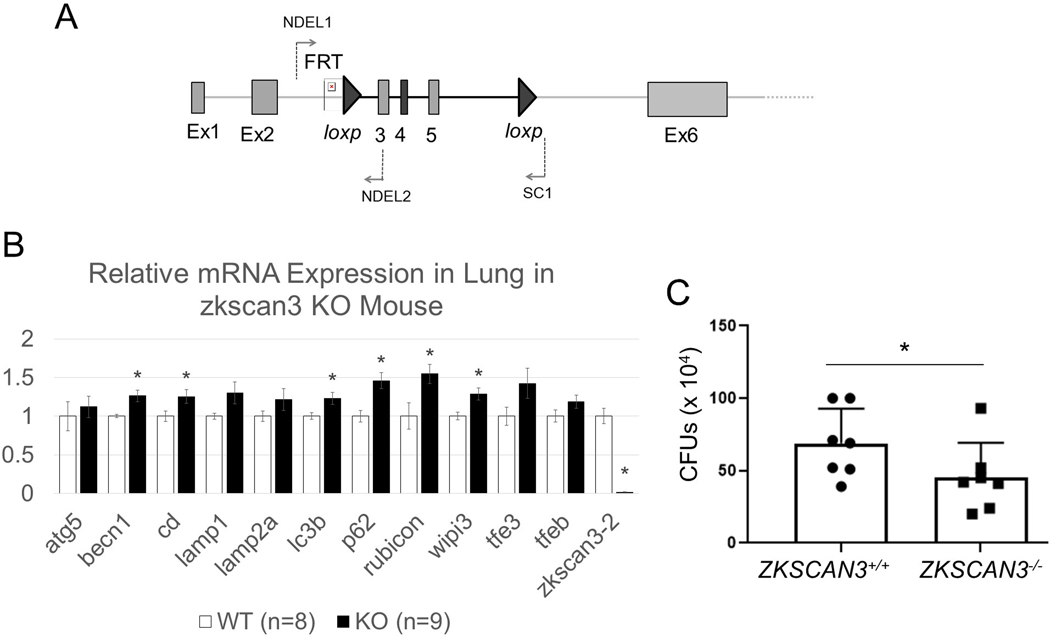
ZKSCAN3 deficiency improves bacterial killing in murine lungs.
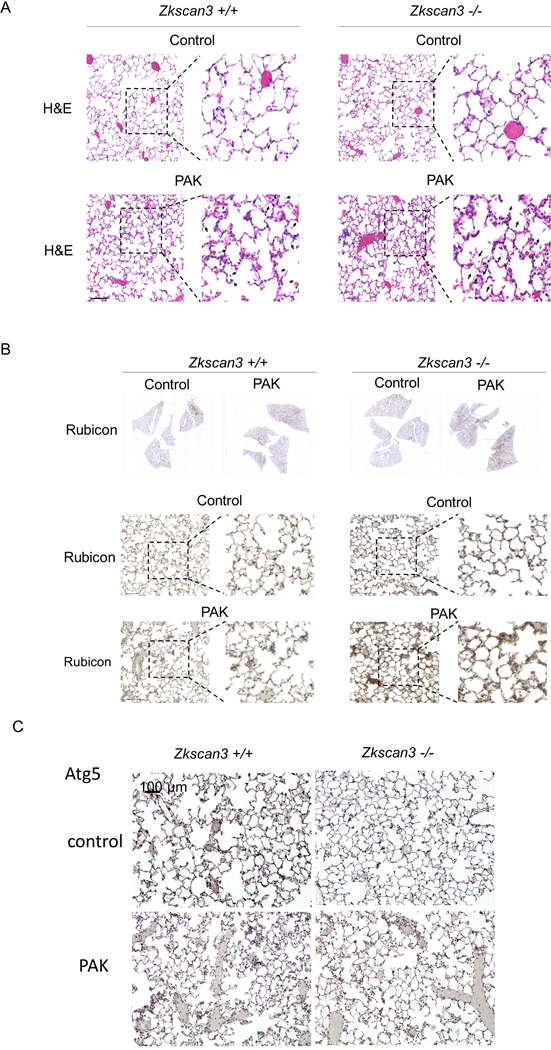
To test if ZKSCAN3 deficiency affects bacterial killing, we generated Zkscan3f/f mice through homologous recombination, followed by generating ZKSCAN3 deficient mice by Cre-Lox recombination through cross breeding Zkscan3f/f and β-actin-Cre (Fig. 3A). Quantitative analysis of Zkscan3 mRNA (RT-PCR) confirmed the knockout, as compared to wild-type mice. In lung homogenates, Zkscan3−/− mice exhibited increased expression of Becn1, Ctsd, Lc3b, Sqstm1, Rubcn and Wipi3, compared to wild-type mice (Fig. 3B). The increased levels of mediators for autophagy, phagosome and lysosome biogenesis were associated with improved killing of PAK in lungs of Zkscan3−/− mice (Fig. 3C). Of note, while PAK induced a similar flux of immune cells in lungs of wild-type and Zkscan3−/− mice, enhanced killing of bacteria was accompanied with a substantial increase of RUBICON in Zkscan3−/− mice (Fig. 4A and B). Moreover, the ATG5 immunostaining is decreased after PAK instillation in wild-type mice, but not in Zkscan3−/− mice (Fig. 4C).
Figure 3.
ZKSCAN3 deficiency improved bacterial killing in lung of mice. (A) Zkscan3 genomic structure with indicated exons, FRT/loxP and loxP sites, as well as the primers used for genotyping. (B) The mRNA profile for indicated autophagy, phagolysosomal and lysosomal components in lungs of wild-type and ZKSCAN3−/− mice. Data normalized to the level of actin expression. Mean ± sem, n = 9, *p < 0.05 (Student’s t-test). (C) The colony forming units (CFUs) from lungs of wild-type and ZKSCAN3 deficient mice that were exposed to PAK (2.5 ×107; i.t.) for 4 hours. Mean ± sem, n = 7 per each groups of mice, *p < 0.05 (Student’s t-test).
Figure 4.
(A) Images of H&E stained lung sections from wild-type or Zkscan3−/− mice that were subjected with or without PAK (2.5 ×107; i.t.) for 4 hours. Black arrows indicate spaces filled with a mixed mononuclear/neutrophilic infiltrates and cellular debris. Alveolar walls are also thickened, and the septa are edematous (blue arrows). (B and C) Images of lung sections with immunostaining for (B) Rubicon or (C) ATG5 in wild-type or ZKSCAN3 deficient mice, including with or without exposure to PAK.
The effect of post-sepsis immunosuppression on phagolysosomes, lysosomes and ZKSCAN3 in murine lungs.
Previous studies have shown that similar to critically-ill patients, post-septic mice have decreased capacity for bacterial killing 40,43. Given that P. aeruginosa-induced pneumonia is associated with a robust decrease in autophagy and lysosomes in lungs of infected mice, we tested if this is relevant in the mice that survive sepsis, but developed immunosuppression. Of note, we have recently demonstrated that mice developed immunosuppression following CLP-induced intra-abdominal sepsis vs. sham 19. This was confirmed by measurement of colony forming units (CFUs) from lung homogenates. In post-sepsis mice, i.e., CLP-induced immunosuppressed mice, the LC3-II/β-actin levels was modestly increased, but without a significant alteration in the LC3-II/LC3-I ratio or lysosomal membrane protein LAMP1 (Supplementary Fig. 1A–C). These findings indicate that in contrast to phagolysosome and lysosome depletion after severe infection of lungs with P. aeruginosa, intra-abdominal infections (seven days post-CLP) had a modest impact on the amounts of phagolysosomes and lysosomes in lungs of mice that survived sepsis. However, the immunosuppressive phenotype is consistent with a robust accumulation of ZKSCAN3, while levels of TFEB and TFE3 remain unaltered (Supplementary Fig. 2A–C). Notably, ZKSCAN3 accumulation was likely linked to its posttranslational regulation, as similar levels of Zkscan3 mRNA, as well as Tfeb and Tfe3 mRNAs, were found in lungs of control and post-sepsis mice (Supplementary Fig. 3A–C).
The expression levels (mRNA) for autophagy and mitochondrial dynamics pathways, including Lc3, Wipi2, Mcoln1, p62, Becn1, Drp1, Prkn, and Pgc1α were not affected in control vs. post-CLP groups of mice (Supplementary Fig. 3D–K). However, lysosomal Lamp1, Lamp2A and Ctsd expression was suppressed, consistent with ZKSCAN3 accumulation in lungs of post-sepsis mice (Supplementary Fig. 4A–C). Interestingly, genes encoding enzymes detoxifying mitochondrial superoxide and cytosolic hydrogen peroxide, Sod2 and Catalase respectively, were also diminished in mice that survived sepsis (Supplementary Fig. 4D–F). Further analysis indicates that the expression levels of Nqo1 and Gclc, target genes of the NRF2 pathway, were similar between tested groups of mice (Supplementary Fig. 4G–H). In regards to metabolic pathways, we observed that expression of genes encoding enzymes for the pentose phosphate pathway and glutathione synthesis (G6pd, Gclm and Gsr) were significantly increased (Supplementary Fig. 4T-V). These findings demonstrate that in an experimental model of sepsis-induced immunosuppression, ZKSCAN3 accumulation is associated with suppressed expression of several ROS detoxifying enzymes and likely affects the pentose phosphate pathway and glutathione synthesis.
Discussion
This is the first study, to our knowledge, to demonstrate an adverse impact of ZKSCAN3 accumulation in experimental models of bacterial lung infections and its accumulation in bacterial lung infection and post-sepsis immunosuppression. Specifically, we found that ZKSCAN3 accumulates in the lungs after severe bacterial pneumonia and also in mice that survived intra-abdominal bacterial sepsis, but developed immunosuppression. ZKSCAN3 deficient mice have enhanced expression of genes encoding autophagy and lysosomal proteins, as well as accumulation of RUBICON protein compared to wildtype mice both with and without exposure to bacterial pneumonia. Importantly. ZKSCAN3−/− mice improved bacterial killing, as evidenced in mice with intratracheal instillation of P. aeruginosa. These findings are consistent with prior studies that Rubicon is essential for LC3 lipidation on phagolysosomes and that ZKSCAN3 is a master transcription repressor for the expression of autophagy and lysosome genes 44–46. Indeed, ZKSCAN3 regulates mRNA expression of Becn-1, Ctsd, Lc3b, Sqstm1, Rubcn and Wipi3. However, we found that Atg5, Lamp1, Lamp2a, Tfe3 or TFEB mRNA levels were not affected in ZKSCAN3−/− mice. Moreover, in response to severe bacterial infection, there is a robust increase of ZKSCAN3 protein in lungs, while the amounts of TFEB, TFE3, LAMP1, and LC3II proteins were reduced.
ZKSCAN3 accumulation is also relevant in mice with sepsis-induced immunosuppression. In this setting, immunosuppression is associated with a robust increase of ZKSCAN3, although mRNA levels was unaltered, as compared to control groups of mice. The mechanism of ZKSKAN3 accumulation remains to be determined, although it is likely linked to posttranslational events. Many transcriptional factors undergo post-translational modifications, including phosphorylation of TFEB that affects its retention and transcriptional function in the nucleus 47.
Decline or loss of autophagy has a tremendous impact on lung function during severe infections and poses high risk of secondary bacterial infections due to immunosuppression developed among survivors of severe sepsis or trauma/hemorrhage 48. Moreover, autophagy is not only limited to neutralization of microbial pathogens, but also affects the severity of lung injury in sterile conditions 49. Lung fibrosis is also known to be accompanied by a loss of autophagy/mitophagy that prevents extracellular matrix clearance by phagocytes and parenchymal cells 50,51. In addition to lowering endoplasmic reticulum stress-induced lung injury, autophagy controls neutralization of damaged/depolarized mitochondria (mitophagy); thus preventing the release of harmful mitochondrial danger-associated molecular patterns (DAMPs) 52. It is possible that ZKSCAN3-dependent depletion of phagolysosomes and lysosomes attenuates recovery from lung injury, thereby leading to post-sepsis complications, including organ fibrosis.
While our studies demonstrate the importance of ZKSCAN3, it is possible that other mechanisms affect phagolysosome and lysosome biogenesis. For example, phagolysosome biogenesis can be enhanced by transcriptional mechanisms that involves the forkhead box O (FOXO) family 53,54. Recent studies have shown that microRNAs (miRNAs) and histone modifications have a significant impact on autophagy 55–58. Of note, ZKSCAN3 may also affect expression of genes that are not related to autophagy, including cyclin D2 and integrin β4 59,60. Although our studies are primarily focused on the ZKSCAN3/autophagy axis, we found that immunosuppression and accumulation of ZKSCAN3 affect expression patterns for SOD2, Cat, G6PD, Gclm, and Gsr. Further studies should consider ZKSCAN3 transcriptional function and regulation in individual cell populations, and importantly, to confirm if similar mechanism/s are operational in the clinical setting of sepsis and immunosuppression.
Taken together, our studies revealed that ZKSCAN3 is associated with depletion of phagolysosomes and lysosomes in lungs of mice with severe infections and during post-sepsis immunosuppression. We suggest that ZKSCAN3 inhibition and/or stimulation of alternative pathways for lysosome biogenesis are important therapeutic targets in sepsis and post-sepsis complications.
Supplementary Material
Acknowledgement.
This study is funded by: US Department of Defense grant W81XWH-17–1-0577 (JWZ);NIH 1R01HL139617–01 (JWZ); Nathan Shock Center NIH AG050886 (VDU, JZ).
Footnotes
Data Availability. All data is available.
Conflict of interest. Authors declare no conflict of interest.
References
- 1.Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR, Vincent JL. Sepsis and septic shock. Nat Rev Dis Primers 2: 16045;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med 369: 2063;2013. [DOI] [PubMed] [Google Scholar]
- 3.Delano MJ, Ward PA. Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J Clin Invest 126: 23–31;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutschman CS, Tracey KJ. Sepsis: current dogma and new perspectives. Immunity 40: 463–475;2014. [DOI] [PubMed] [Google Scholar]
- 5.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 306: 2594–2605;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward PA. Immunosuppression in sepsis. JAMA 306: 2618–2619;2011. [DOI] [PubMed] [Google Scholar]
- 7.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 13: 862–874;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest 116: 984–995;2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Bhatia M, Zhu YZ, Zhu YC, Ramnath RD, Wang ZJ et al. Hydrogen sulfide is a novel mediator of lipopolysaccharide-induced inflammation in the mouse. FASEB J 19: 1196–1198;2005. [DOI] [PubMed] [Google Scholar]
- 10.Bhatia M, Wong FL, Fu D, Lau HY, Moochhala SM, Moore PK. Role of hydrogen sulfide in acute pancreatitis and associated lung injury. FASEB J 19: 623–625;2005. [DOI] [PubMed] [Google Scholar]
- 11.Giordano S, Darley-Usmar V, Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol 2: 82–90;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryter SW, Choi AM. Autophagy in lung disease pathogenesis and therapeutics. Redox biology 4: 215–225;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 368: 1845–1846;2013. [DOI] [PubMed] [Google Scholar]
- 14.Mizumura K, Cloonan SM, Haspel JA, Choi AMK. The emerging importance of autophagy in pulmonary diseases. Chest 142: 1289–1299;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science 340: 697–701;2013. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists-Mechanisms and experimental approaches. Redox Biol 4C: 242–259;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li MX, Sun XM, Zhang LZ, Wang J, Huang YZ, Sun YJ et al. A novel c.−274C>G polymorphism in bovine SIRT1 gene contributes to diminished promoter activity and is associated with increased body size. Animal genetics 44: 584–587;2013. [DOI] [PubMed] [Google Scholar]
- 18.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ 20: 31–42;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bone NB, Becker EJ Jr., Husain M, Jiang S, Zmijewska AA, Park DW et al. AMPK activates Parkin independent autophagy and improves post sepsis immune defense against secondary bacterial lung infections. Sci Rep 11: 12387;2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 15: 65–74;2014. [DOI] [PubMed] [Google Scholar]
- 21.Wani WY, Ouyang X, Benavides GA, Redmann M, Cofield S, Shacka JJ et al. O-GlcNAc regulation of autophagy and α-synuclein homeostasis; implications for Parkinson’s disease. Molecular Brain;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodson M, Wani WY, Redmann M, Benavides GA, Johnson M, Ouyang X et al. Regulation of autophagy, mitochondrial dynamics and cellular bioenergetics by 4-hydroxynonenal in primary neurons. Autophagy;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wani WY, Chatham JC, Darley-Usmar V, McMahon LL, Zhang J. O-GlcNAcylation and neurodegeneration. Brain Res Bull;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J. Regulation of autophagy by protein post-translational modification. Lab Invest 95: 14–25;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S et al. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sardiello M, Palmieri M, di RA, Medina DL, Valenza M, Gennarino VA et al. A gene network regulating lysosomal biogenesis and function. Science 325: 473–477;2009. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Jing Y, Qin Y, Hunsucker S, Meng H, Sui J et al. The zinc finger transcription factor ZKSCAN3 promotes prostate cancer cell migration. Int J Biochem Cell Biol 44: 1166–1173;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell 50: 16–28;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan H, Yan Y, Liu C, Finkel T. The role of ZKSCAN3 in the transcriptional regulation of autophagy. Autophagy 13: 1235–1238;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saftig P, Haas A. Turn up the lysosome. Nat Cell Biol 18: 1025–1027;2016. [DOI] [PubMed] [Google Scholar]
- 31.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 15: 65–74;2014. [DOI] [PubMed] [Google Scholar]
- 32.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell 50: 16–28;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Song Y, Quach C, Guo H, Jang GB, Maazi H et al. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat Commun 10: 1693;2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi Y, Xu H, Wang F, Chen X, Shan Z, Sun Y et al. ZKSCAN3 promotes breast cancer cell proliferation, migration and invasion. Biochem Biophys Res Commun 503: 2583–2589;2018. [DOI] [PubMed] [Google Scholar]
- 35.Kawahara T, Inoue S, Ide H, Kashiwagi E, Ohtake S, Mizushima T et al. ZKSCAN3 promotes bladder cancer cell proliferation, migration, and invasion. Oncotarget 7: 53599–53610;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady OA, Martina JA, Puertollano R. Emerging roles for TFEB in the immune response and inflammation. Autophagy 14: 181–189;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurundkar D, Kurundkar AR, Bone NB, Becker EJ Jr., Liu W, Chacko B et al. SIRT3 diminishes inflammation and mitigates endotoxin-induced acute lung injury. JCI Insight 4;2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carles M, Wagener BM, Lafargue M, Roux J, Iles K, Liu D et al. Heat-shock response increases lung injury caused by Pseudomonas aeruginosa via an interleukin-10-dependent mechanism in mice. Anesthesiology 120: 1450–1462;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bone NB, Liu Z, Pittet JF, Zmijewski JW. Frontline Science: D1 dopaminergic receptor signaling activates the AMPK-bioenergetic pathway in macrophages and alveolar epithelial cells and reduces endotoxin-induced ALI. J Leukoc Biol 101: 357–365;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Bone N, Jiang S, Park DW, Tadie JM, Deshane J et al. AMP-activated protein kinase and Glycogen Synthase Kinase 3beta modulate the severity of sepsis-induced lung injury. Mol Med;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregoire M, Tadie JM, Uhel F, Gacouin A, Piau C, Bone N et al. Frontline Science: HMGB1 induces neutrophil dysfunction in experimental sepsis and in patients who survive septic shock. J Leukoc Biol 101: 1281–1287;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Settembre C, Di MC, Polito VA, Garcia AM, Vetrini F, Erdin S et al. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park DW, Zmijewski JW. Mitochondrial Dysfunction and Immune Cell Metabolism in Sepsis. Infection & chemotherapy 49: 10–21;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong SW, Sil P, Martinez J. Rubicon: LC3-associated phagocytosis and beyond. FEBS J 285: 1379–1388;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17: 893–906;2015. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Herb M, Gluschko A, Schramm M. LC3-associated phagocytosis - The highway to hell for phagocytosed microbes. Semin Cell Dev Biol 101: 68–76;2020. [DOI] [PubMed] [Google Scholar]
- 47.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14: 283–296;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol 19: 170–183;2019. [DOI] [PubMed] [Google Scholar]
- 49.Maciel M, Hernandez-Barrientos D, Herrera I, Selman M, Pardo A, Cabrera S. Impaired autophagic activity and ATG4B deficiency are associated with increased endoplasmic reticulum stress-induced lung injury. Aging (Albany NY) 10: 2098–2112;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest 125: 521–538;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rangarajan S, Bone NB, Zmijewska AA, Jiang S, Park DW, Bernard K et al. Metformin reverses established lung fibrosis in a bleomycin model. Nat Med 24:1121–1127;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Youle RJ. Mitochondria-Striking a balance between host and endosymbiont. Science 365;2019. [DOI] [PubMed] [Google Scholar]
- 53.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6: 458–471;2007. [DOI] [PubMed] [Google Scholar]
- 54.Im J, Hergert P, Nho RS. Reduced FoxO3a expression causes low autophagy in idiopathic pulmonary fibrosis fibroblasts on collagen matrices. Am J Physiol Lung Cell Mol Physiol 309: L552–561;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J et al. Function and molecular mechanism of acetylation in autophagy regulation. Science 336: 474–477;2012. [DOI] [PubMed] [Google Scholar]
- 56.Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-DeltaNp63alpha-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell cycle 11: 1247–1259;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bryant A, Palma CA, Jayaswal V, Yang YW, Lutherborrow M, Ma DD. miR-10a is aberrantly overexpressed in Nucleophosmin1 mutated acute myeloid leukaemia and its suppression induces cell death. Mol Cancer 11: 8;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu H, Wang F, Hu S, Yin C, Li X, Zhao S et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal 24: 2179–2186;2012. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, Wang H, Kornblau SM, Graber DA, Zhang N, Matthews JA et al. Evidence of a role for the novel zinc-finger transcription factor ZKSCAN3 in modulating Cyclin D2 expression in multiple myeloma. Oncogene 30: 1329–1340;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L, Zhang L, Wu Q, Boyd DD. Unbiased screening for transcriptional targets of ZKSCAN3 identifies integrin beta 4 and vascular endothelial growth factor as downstream targets. J Biol Chem 283: 35295–35304;2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.