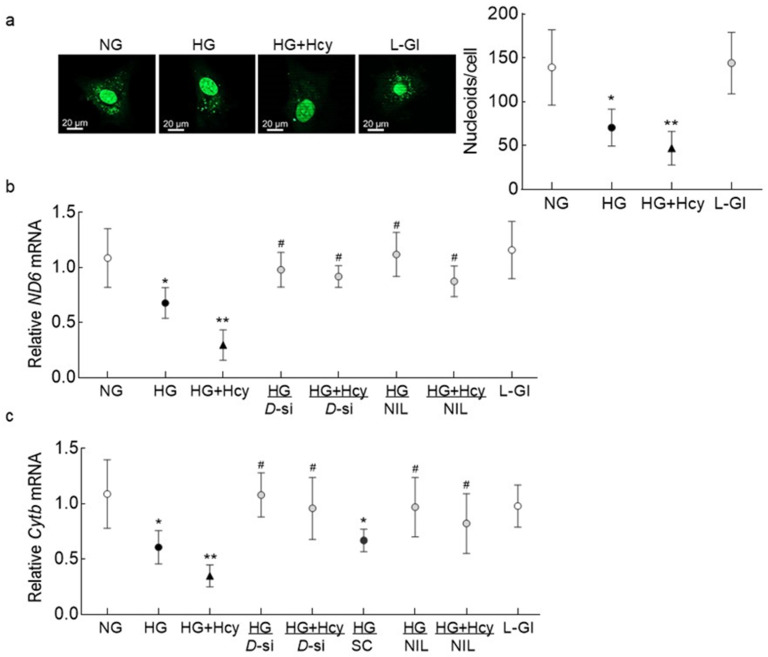
Figure 5.
Effect of homocysteine on mitochondrial nucleoids and transcription of mtDNA. (a) Nucleoids were quantified using SYBR DNA gel stain, and the images were acquired using a 40× objective. The accompanying graph has values represented as mean ± SD obtained from 30–40 cells in each group. Gene transcripts of mtDNA-encoded (b) ND6 and (c) Cytb were quantified by qRT-PCR using β-actin as a housekeeping gene. The values are represented as mean ± SD obtained from 3–4 different cell preparations with each measurement made in triplicate: NG and HG, cells in normal and high glucose, respectively; HG + Hcy, cells in high glucose and homocysteine; HG/D-si and HG + Hcy/D-si, Drp1-siRNA transfected cells in high glucose or high glucose + homocysteine, respectively; HG/SC, nontargeting scrambled RNA transfected cells in high glucose; HG/NIL and HG + Hcy/NIL, cells in the presence of NIL in high glucose or high glucose + homocysteine, respectively; L-Gl, 20 mM L-glucose; * and ** p < 0.05 vs. NG and HG, respectively; # p > 0.05 vs. NG.