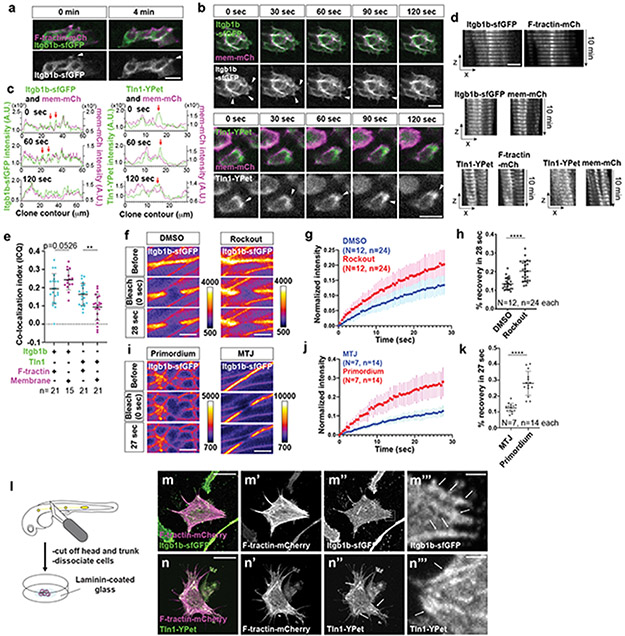
Extended Data Fig. 4. Integrin-β1b and Talin1 dynamics in cells of the primordium.
a, Localization of Itgb1b-sfGFP and F-tractin-mCherry at the apical side of superficial cells in the primordium. The images are single optical slices. Arrowheads indicate Itgb1b-sfGFP clustering. Scale bar = 10 μm. b, Localization of Itgb1b-sfGFP (top) and Tln1-YPet (bottom) with membrane-mCherry at the basal sides of cells in clones in the primordium imaged over time taken from Video 3. The images are single optical slices. Arrowheads indicate Itgb1b-sfGFP and Tln1-YPet clustering. Scale bar = 10 μm. c, Intensity profiles of Itgb1b-sfGFP (left) and Tln1-YPet (right) together with membrane-tethered mCherry along the contours of clones at indicated times taken from Video 3. Arrows indicate Itgb1b-sfGFP and Tln1-YPet clusters that do not coincide with membrane-tethered mCherry clustering. Representative profile of 5 or more imaged cells. d, Montage of 10 consecutive images of the basal sides of the clones. The images are single transverse sections from a time lapse video. Scale bar = 10 μm. e, Quantification of co-localization of Itgb1b-sfGFP and Tln1-YPet with F-tractin-mCherry and membrane tethered mCherry. Li’s ICQ co-localization indices of 0.5 and −0.5 indicate perfectly co-localized and perfectly anti-co-localized signals, respectively. n = number of cells. Data points, means, and SD are indicated. Three data points were analyzed from the same embryo. **: p=0.0015 (two-tailed t-test). f, Images from time-lapse video after photo-bleaching of Itgb1b-sfGFP at the myotendinous junction of embryos treated with DMSO or 50 μM Rockout. GFP intensities are pseudo-colored as a heat map. Scale bars = 10 μm. g, Graph of Itgb1b-sfGFP fluorescence intensity over time before and after photo-bleaching in embryos treated with DMSO or 50 μM Rockout. The fluorescence intensities are normalized to the minimal intensities after photo-bleaching. Dots indicate mean intensities and error bars are SD. n = number of experiments, N = number of embryos. h, Plot of the percent recovery of Itgb1b-sfGFP fluorescence intensity at 28 sec after photo-bleaching in embryos treated with DMSO or 50 μM Rockout. Data points, means, and SD are indicated. ****: p<0.0001 (two-tailed Welch’s t-test). n = number of experiments (used for statistical test), N = number of embryos. i, Images from time-lapse video after photo-bleaching of Itgb1b-sfGFP in the primordium (left) and at the myotendinous junction (right). Fluorescence intensities are pseudo-colored as a heat map. Scale bars = 10 μm. j, Graph of Itgb1b-sfGFP fluorescence intensity over time before and after photo-bleaching in the primordium and at the myotendinous junction. The fluorescence intensities are normalized to the minimal intensities after photo-bleaching. Dots indicate mean intensities and error bars are SD. n = number of experiments, N = number of embryos. k, Plot of the percent recovery of Itgb1b-sfGFP fluorescence intensity at 27 sec after photo-bleaching in the primordium and at the myotendinous junction. n = number of experiments (used for statistical test), N = number of embryos. Data points, means, and SD are indicated. ****: p<0.0001 (two-tailed Welch’s t-test). l, Experimental design to culture primordium cells. m, Antibody staining against Itgb1b-GFP and F-tractin-mCherry on cultured primordium cells. Arrowheads indicate actin stress fibers (m’) with Itgb1b-GFP clusters (arrows in m”) in the cell center and in protrusions (m”’). Scale bars = 20 μm (m-m”) and 1 μm (m”’). n, Antibody staining against Tln1-YPet and F-tractin-mCherry on cultured primordium cells. Arrowheads indicate actin stress fibers (n’) with Tln1-YPet clusters (arrows in n”) in the cell center and in protrusions (n”’). Scale bars = 20 μm (n-n”) and 1 μm (n”’). Images are max-projected z-stacks. Close-ups (right panels) are magnifications of the regions indicated by dotted squares in the middle panels.