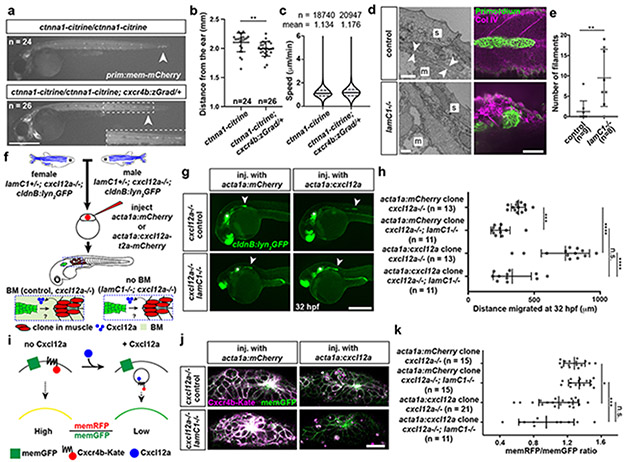
Fig. 2. Primordium migration requires an intact basement membrane.
a, Control and Ctnna1-depleted primordia (arrowheads) in 48 hpf embryos. Scale bar = 0.5 mm. Close-up of region indicated by a dashed square. b, Quantification of the migration distance for primordia shown in a. Individual data points, means and SD are indicated. **: p=0.0013 (two-tailed Mann-Whitney test). c, Speed of Ctnna1-depleted primordium cells. Solid line = median, dashed line = quartile. n = cell speeds from more than 7 primordia with each more than 100 cells). d, Left. TEM images of the ultrastructure of the BM between the skin (s) and the muscle (m) in control (n = 2) and lamC1−/− embryos (n = 1). White arrows indicate the BM. Scale bar = 2 μm. Right. Antibody staining against Collagen IV in control and lamC1−/− embryos. Scale bar = 50 μm. e, Quantification of Collagen IV filaments in control and lamC1−/− embryos. Individual data points, means and SD are indicated. **: p=0.0056 (two-tailed Mann-Whitney test). f, Strategy to express Cxcl12a in a few muscle cells in lamC1−/− embryos and siblings. g, Images of the migrating primordium in cxcl12a−/− and cxcl12a−/−; lamC1−/− 32 hpf embryos with clones in the trunk muscle that express mCherry (not shown) or Cxcl12a together with mCherry (not shown). Asterisks indicate the ear and arrowheads the primordium. Scale bar = 0.5 mm. h, Quantification of the distance migrated by the primordium in the indicated experimental conditions at 32 hpf. Individual data points, means and SD are indicated. ***: p=0.0002, ****: p<0.0001, n.s: p=0.0879 (two-tailed Mann-Whitney test). i, Principle of the Cxcl12a sensor. j, Images of the Cxcl12a sensor in primordia of cxcl12a−/− and cxcl12a−/−; lamC1−/− live embryos with clones in the muscle of the trunk that express mCherry or Cxcl12a. Scale bar = 20 μm. k, Quantification of the Cxcr4b-Kate-to-memGFP ratio in the primordia of embryos shown in j. Individual data points, means and SD are indicated. *: p=0.038, ***: p=0.0001, n.s: p=0.1349 (one way ANOVA followed by Holm-Sidak’s multiple comparison test). Note, controls are lamc1+/+ and lamc1−/+ embryos. For a, b, e, h, k, n = number of embryos.