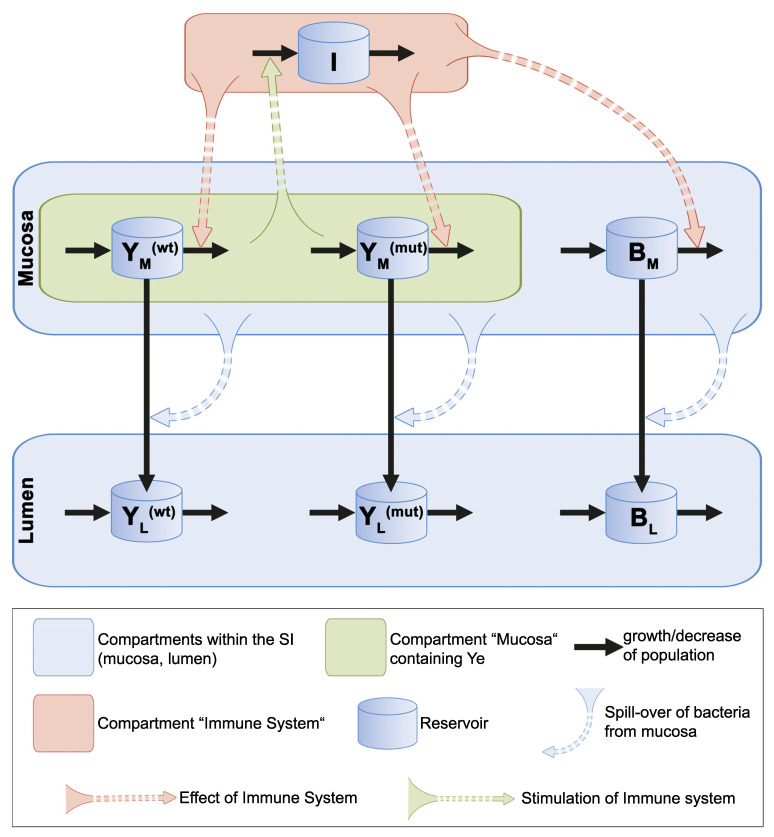
Figure 3.
Schematic graphical depiction of the model composition and interaction networks. The model calculates population dynamics of the Ye wt (YL(wt); YM(wt)) and mutant strains (YL(mut), YM(mut)), as well as of commensal bacteria (BL; BM) at two different sites of the small intestine (SI), the luminal site and the extra-luminal mucosal site (“mucosa”; “lumen”). Additionally, it includes an abstract immune response with a distinct immune cell population (I). Bacterial and immune cell populations are illustrated as reservoirs. Individual growth rates determine the growth of bacterial populations. The decrease in populations is caused by intestinal peristaltic movement in the lumen and by immune killing in the mucosa. In addition, movement of bacteria from the mucosal compartment to the luminal compartment takes place. Upon entry of Ye wt or mutant strains to the mucosal compartment, they stimulate an immune response, which reciprocally affects all Ye and commensal populations within this compartment. The Ye wt strain, equipped with immune evasion factors, is less affected by the immune response than the Ye mutant strain, whereas both are more resistant than the commensal bacterial population (BM). Replicating populations that exceed the limited capacity of the mucosa drain into the lumen and, thereby, feed luminal populations. As a result of these bacterial population dynamics in the lumen, the model output is the calculated CFU of the bacteria ending up in feces. These curves are equivalent to experimental CFU data generated from the feces of orally infected mice.