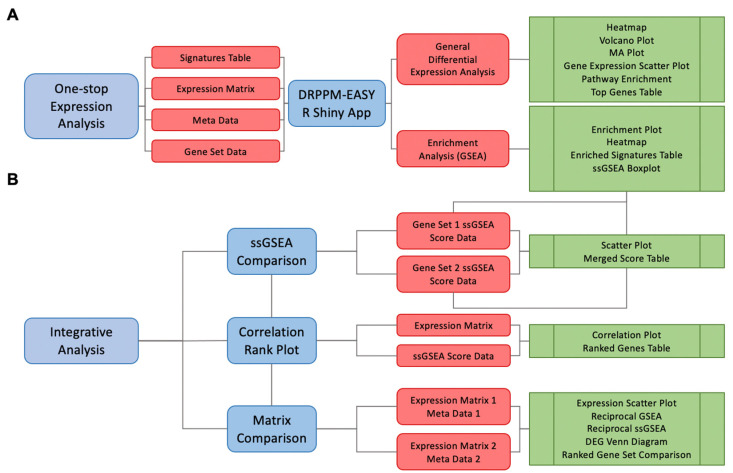
Figure 1.
DRPPM-EASY expression analysis pipeline. (A) Schematic workflow of DRPPM-EASY. The pipeline takes in input files of an expression matrix, a sample meta-file specifying sample grouping, and a gene set database for GSEA. A GSEA enriched signature table is generated as a preprocessing step, which is used as input to the R Shiny app. The app generates two modes of exploring the data: (1) general differential gene expression analysis and (2) gene set enrichment analysis. The result from the analysis can be downloaded as output tables. (B) Schematic of the integrative analysis with three major features for pathway signature comparison. The app has three modes of integrative analysis: (1) scatter plot mode, (2) correlation plot mode, and (3) paired multi-omics analysis.