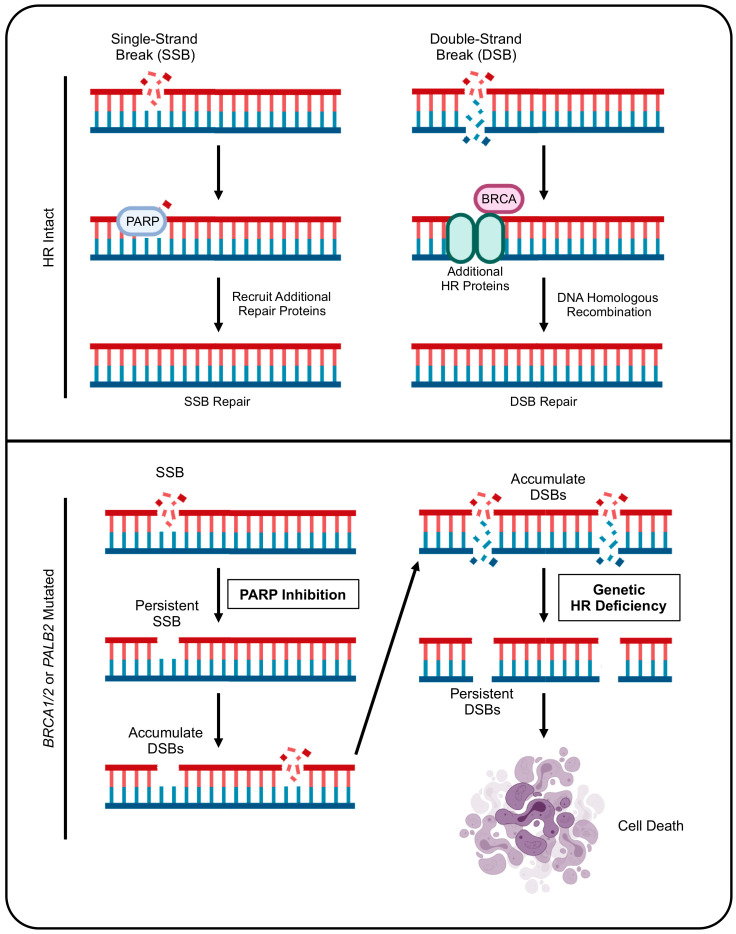
Figure 1.
Molecular basis for the efficacy of PARP inhibition in HR-deficient tumor cells. In homologous recombination (HR) proficient PDAC cells, single-strand DNA breaks (SSBs) lead to the rapid synthesis and recruitment of the DNA damage sensor poly (ADP-ribose) polymerase (PARP). In brief, once the PARP enzyme recognizes the DNA breaks via its DNA-binding domain, PARP facilitates base excision repair by acting as a scaffold to recruit additional DNA repair proteins including PNKP, APTX, and LIG3. These additional repair proteins process the SSB, and the gap in the DNA strand is then filled by DNA polymerases and ligated by LIG1. For more severe double-strand breaks (DSBs), protein kinases including ATM and ATR act as damage sensors, driving the recruitment of BRCA proteins to the site of DNA damage. BRCA proteins, assisted by others including BARD1 and BRIP1, organize the assembly of several other DNA repair proteins. This culminates in RAD51 loading, strand invasion, DNA synthesis, and HR-mediated DNA repair to maintain genomic integrity. PDAC cells with deleterious mutations to BRCA or PALB2 are deficient in HR repair, and unable to accommodate DSBs. Therefore, by disrupting the ability of these cells to repair SSB repair using PARP inhibitions, these cells accumulate DSBs, resulting in DNA fragmentation and programmed cell death.