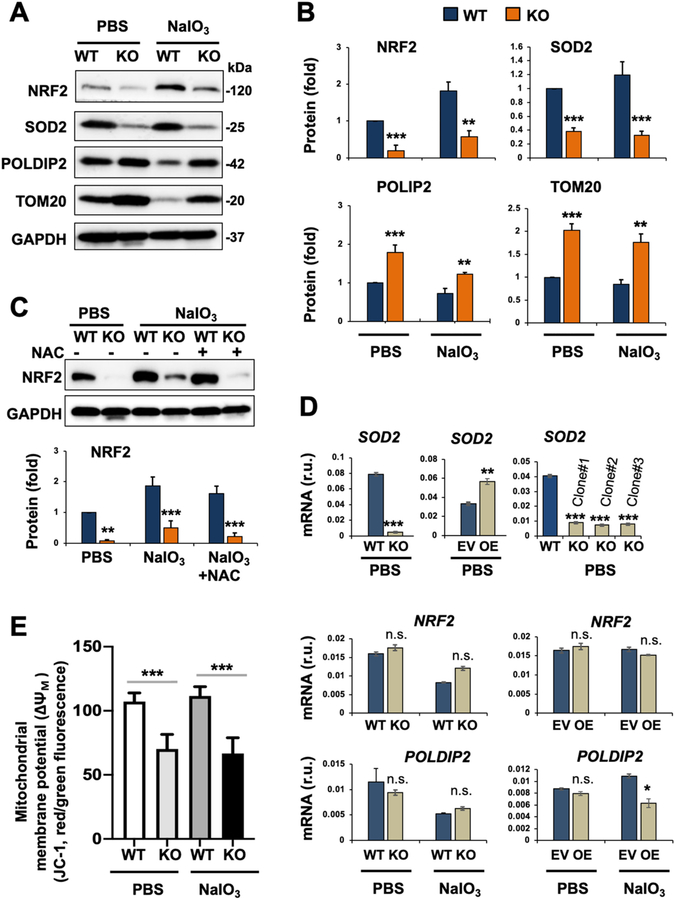
Fig. 6. Reduced NRF2 protein and altered mitochondria-residing (or associated) proteins in oxidatively challenged TMEM97 KO (vs WT) ARPE19 cells.
Prior to cell harvest for various assays, ARPE19 cells were treated with PBS or 5 mM NaIO3 for 24 h, as described for Fig. 5. We chose to use the NaIO3 condition of 5 mM/24 h in the remainder experiments of this study because it was sufficient to activate the apoptotic program yet without causing severe cell death so that various intracellular events could still be detected.
A. Representative Western blots.
B. Quantification for A: Mean ± SEM, n = 6 (for NRF2) and n = 3 independent repeat experiments (for other 3 proteins). Statistics: ANOVA followed by Bonferroni test: *P < 0.05, **P < 0.01, ***P < 0.001, KO vs WT. r.u., relative unit.
C. Lack of NAC effect on TMEM97 KO-induced NRF2 protein down-regulation. NAC was added together with NaIO3. Quantification: Mean ± SEM, n = 3 independent repeat experiments (different time than that in A/B). Statistics: ANOVA followed by Bonferroni test: ***P < 0.001 (KO vs WT).
D. qRT-PCR analysis. EV: empty vector; OE: overexpression. Quantification: Mean ± SD, n = triplicates. Statistics: ANOVA followed by Bonferroni test: *P < 0.05, **P < 0.01, ***P < 0.001, KO vs WT or OE vs EV. n.s., not significant. Shown in each plot is one of two similar experiments. Note: Among the three TMEM97 KO single clones, #1 was used for the rest of this study.
E. Mitochondrial membrane potential (ΔΨM). Assay was performed using the JC-1 kit (3 h NaIO3 treatment). Quantification: Mean ± SD, n = triplicates. Statistics: ANOVA followed by Bonferroni test: ***P < 0.001 (KO vs WT).