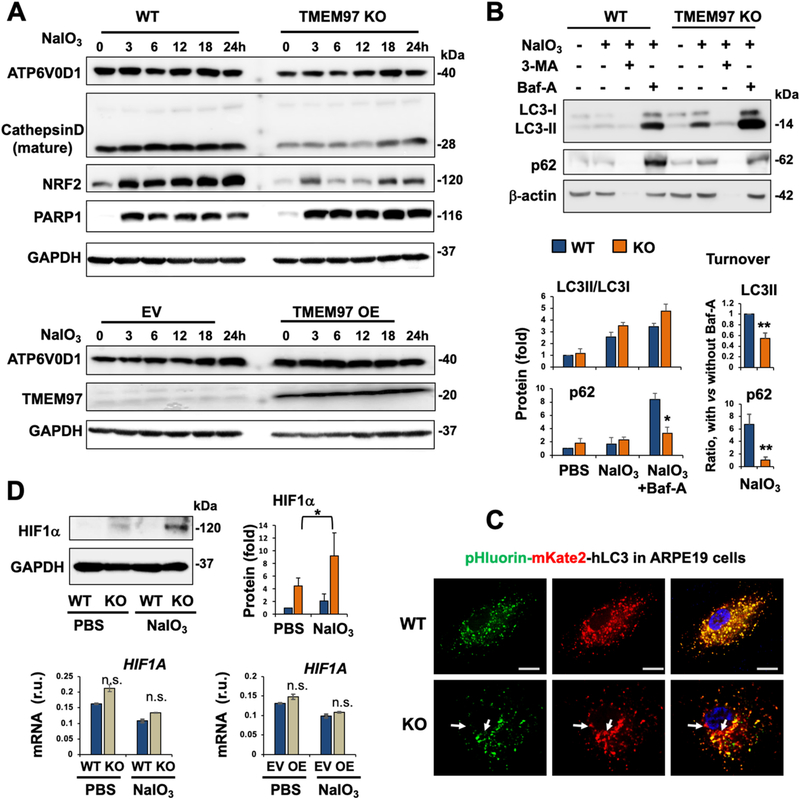
Fig. 7. Altered protein levels or turnover suggestive of compromised lysosomal function in oxidatively challenged TMEM97 KO (vs WT) ARPE19 cells.
Prior to cell harvest for various assays, ARPE19 cells were treated with PBS or 5 mM NaIO3 for 24 h, as described for Fig. 5.
A. Time course of NaIO3 treatment showing reduced ATP6V0D1 and cathepsin-D due to TMEM97 KO. NRF2 and PARP1 were also determined as positive controls for the impact of TMEM97 KO (as seen in Figs. 5 and 6).
B. Autophagosome cargo protein turnover. Cells were treated with PBS or NaIO3, in the presence of vehicle (DMSO) or 10 μM 3-Methyladenine (3-MA, inhibitor of autophagosome biogenesis) or 50 nM bafilomycin-A (Baf-A, inhibitor of lysosomal function such as autophagosome-lysosome fusion). In the process of autophagic flux, LC3II and p62(SQSTM1) are transported to lysosomes via autophagosome-lysosome fusion and thereby degraded. Bafilomycin-A (Baf-A) blocks this lysosomal function. Thus, the difference or delta of the protein levels of p62 (or LC3II) with and without Baf-A measures its turnover or autophagy flux. Note: 3-MA, blocker of autophagosome biogenesis, likely caused severe cell death (see beta-actin bands disappearing).
C. Analysis of autophagosome-lysosome fusion under oxidative stress of NaIO3. Autophagosomes are indicated by pHluorin (GFP variant)-mKate2 (red) in tandem with LC3 (transfected into ARPE19 cells), which appeared as yellow puncta when not located in lysosomes; green (pHluorin) should be quenched inside lysosomes due to their lower pH, causing loss of green fluorescence. More red puncta (e.g. marked by white arrows) in KO vs WT cells suggest that autophagosome-lysosome fusion still occurred and that cargo has accumulated in lysosomes. Scale bar: 20 μm.
D. Increase of HIF1α protein (but not mRNA) in TMEM97 KO vs WT ARPE19 cells. Quantification of Western blots (for B and D): Mean ± SEM, n ≥ 3 independent repeat experiments. Quantification for qRT-PCR: Mean ± SD, n = triplicate. Statistics: ANOVA followed by Bonferroni test: *P < 0.05, **P < 0.01, KO vs WT.