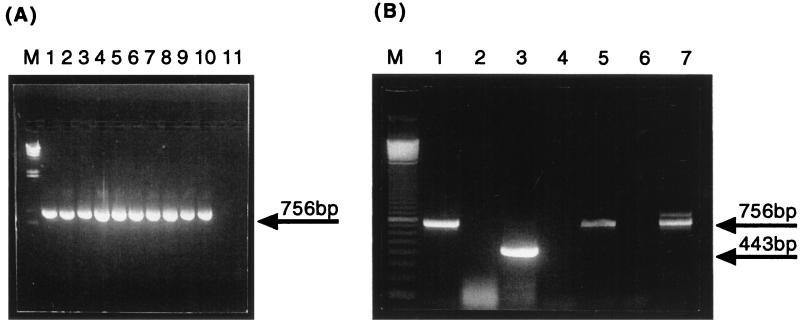
FIG. 1.
PCR and RT-PCR of COX2 genes from total cellular DNA and mtRNA. (A) COX2 was amplified as a 756-bp product from total cellular DNAs of 10 strains of C. glabrata (lanes 1 to 10, respectively) but not from S. cerevisiae total cellular DNA (lane 11). C. glabrata DNA samples correspond to strains 05, 06, 12, 13, 36, 37, 38, 39, 47 and ATCC 90030, respectively (TABLE 1). (B) COX2 was amplified from purified mtRNA of C. glabrata 06 (Table 1) (lanes 5 and 7). The cDNAs were synthesized either from oligo(dT) primers (lane 4) or from the COR primer (lane 7). Negative controls consisted of mock RT reactions with oligo(dT) (lane 4) or COR primer (lane 6) lacking reverse transcriptase to demonstrate that the COX2 product was reverse transcriptase dependent. Amplification of actin cDNA from oligo(dT) was used to control for RNA integrity, RT efficiency, and contamination with nonmitochondrial mRNA (lane 3) with the corresponding mock RT reaction-negative control to exclude the possibility of DNA contamination (lane 2). Amplification of COX2 from total cellular DNA was used as a positive control for PCR (lane 1). The identities of the COX2 and actin cDNA amplicons were checked by sequencing. Lane M, molecular size markers (phage lambda DNA cut with HindIII for panel A and a 100-bp ladder [GIBCO BRL] for panel B).