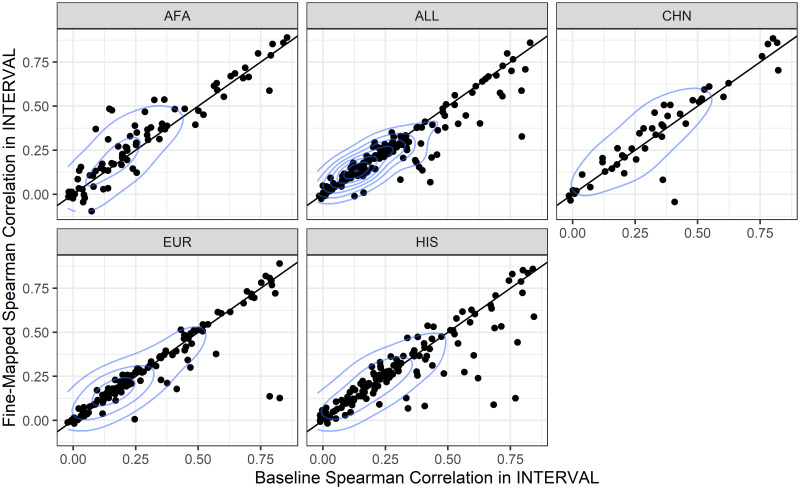
Fig 2. TOPMed MESA protein prediction model performance comparison in the independent INTERVAL population.
Within each training population, the fine-mapped model performance in INTERVAL (y-axis) is compared to the baseline elastic net model performance in INTERVAL (x-axis). Each dot represents a protein that is predicted by both baseline models and fine-mapped models. Performance was measured as the Spearman ρ between the measured protein aptamer level and the predicted protein aptamer level. Fine-mapped models performed better than baseline models in AFA (Wilcoxon signed-rank test, p = 0.0016) and CHN (p = 0.036), were not significantly different in EUR (p = 0.74) and HIS (p = 0.54), and significantly worse in ALL (p = 0.0085). TOPMed MESA populations: AFA, African American; ALL, all populations combined; CHN, Chinese; EUR, European; HIS, Hispanic/Latino.