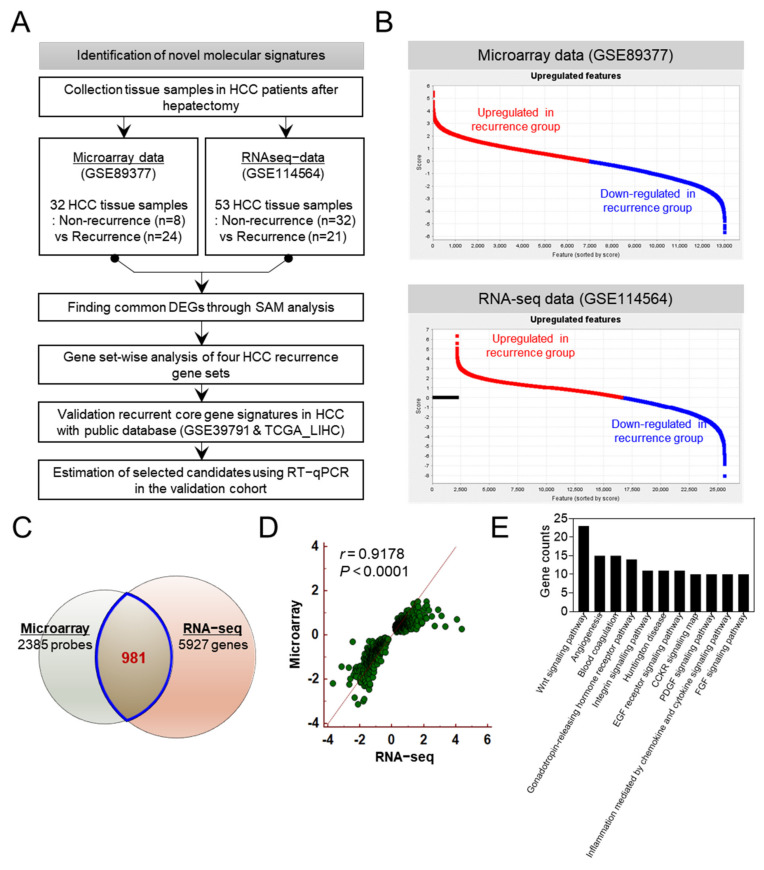
Figure 1.
Integrative analysis of tissue−based microarray data and RNA−seq data to identify novel gene signatures for the recurrence of hepatocellular carcinoma (HCC). (A) Flow chart demonstrating the methodology used to identify gene signatures for predicting the recurrence of hepatocellular carcinoma. (B) Gene expression plot of each dataset displaying upregulated and downregulated genes. (C) Venn diagram analysis to determine common differentially expressed genes in HCC tissues identified using two different datasets. (D) Scatter plot of the correlation between microarray and RNA−seq results for 981 differentially expressed genes (DEGs; r = 0.9178, p < 0.001). (E) The analysis of gene ontology revealed significant enrichment of differentially expressed gene signatures associated with biological processes.