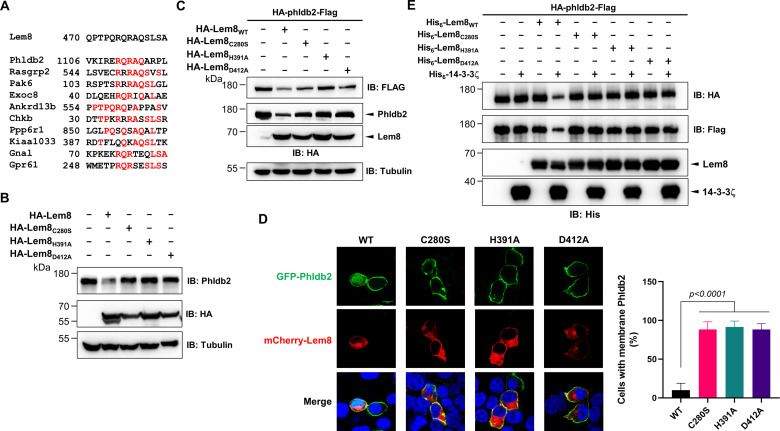
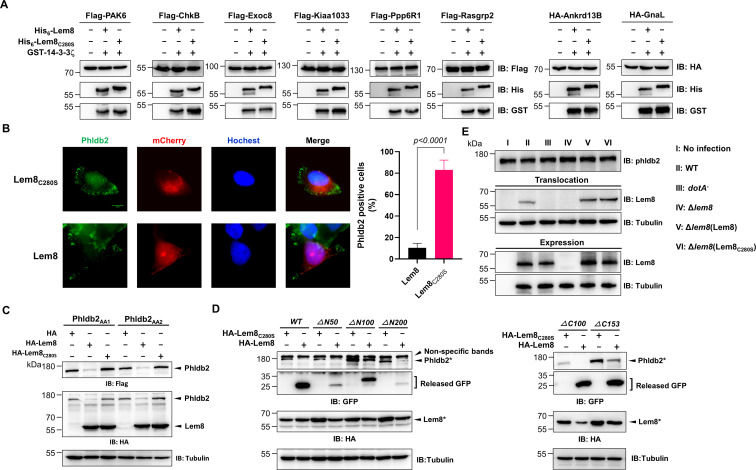
Figure 4. Lem8 cleaves Phldb2 in a manner that requires 14-3-3ζ.
(A) Multiple alignments of the self-cleavage site of Lem8 with potential targets in human cells identified by bioinformatic analysis. Identical residues are highlighted in red. (B) Lem8 reduces the protein levels of endogenous Phldb2 in mammalian cells. Lem8 and the indicated mutants were individually expressed in HEK293T cells by transfection. Twenty-four hours after transfection, the samples were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and detected by immunoblotting with anti-Phldb2 antibodies. Tubulin was used as a loading control. Results shown were one representative from three independent experiments with similar results. (C) Lem8 cleaves exogenous Phldb2 in mammalian cells. HA and Flag tag were fused to the amino and carboxyl end of Phldb2, respectively, and the double tagged protein was coexpressed in HEK293T cells with Lem8 or each of the mutants. Twenty-four hours after transfection, the samples were resolved by SDS–PAGE and probed by a HA-specific antibody and a Flag-specific antibody, respectively. Tubulin was detected as a loading control. Results shown were one representative from three independent experiments with similar results. (D) Lem8 alters the subcellular distribution of GFP fused to Phldb2. GFP was fused to the amino end of Phldb2 and the protein was coexpressed in HEK293T cells with mCherry-Lem8 or each of the mutants. Twenty-four hours after transfection, cells were fixed and nucleus were stained by Hoechst 33,342. The fluorescence Images of GFP (green), mCherry (red), and Hoechst (blue) were acquired with a Zeiss LSM 880 confocal microscope. The percentage of cells with membrane Phldb2 was calculated in Phldb2- and Lem8-positive cells (right panel). Bar, 10 μm. (E) 14-3-3ζ is required for the cleavage of Phldb2 by Lem8. HA-Phldb2-Flag was expressed in HEK293T cells, immunoprecipitated with a Flag-specific antibody, and eluted with 3× Flag peptides. Purified Phldb2 was incubated with His6-Lem8 or each of the mutants in reactions with or without His6-14-3-3ζ. Total proteins of all samples were resolved with SDS–PAGE, and probed by immunoblotting with a HA-specific antibody, a Flag-specific antibody and a His-specific antibody. Results shown were one representative from three independent experiments with similar results.