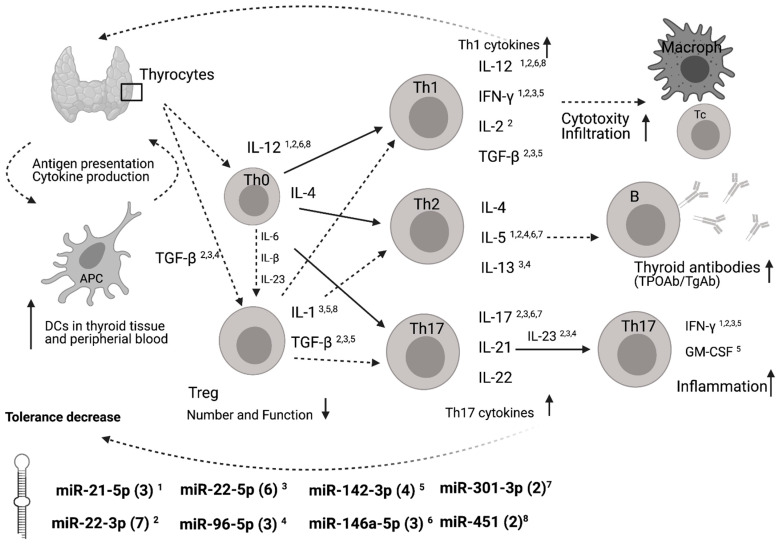
Figure 4.
Summary of the main mechanisms related to autoimmunity of HT and potential interaction sites with differentially expressed miRNAs. Schematic representation of T cells differentiating into specific T cell subsets depending on the cytokines to which they are exposed and their main effects. MiRNA binding site predictions have been annotated by miRWalk database. Number of predicted binding sites are shown in brackets for each miRNA. Predicted binding sites of genes of HT immune-related molecules and/or their receptors are marked by superscript numbers. Adapted from [22]. APC, antigen presenting cell; Th, T helper cell; Macroph, macrophage; DC, dentritic cell; Treg, T-regulatory cells; TPOAb; peroxidase autoantibody; TgAb, thyroglobulin autoantibody, IL, interleukin; IFN-γ, interferon-γ; TGF-β, transforming growth factor β; GM-CSF, granulocyte-macrophage colony-stimulating factor; miR, miRNA.