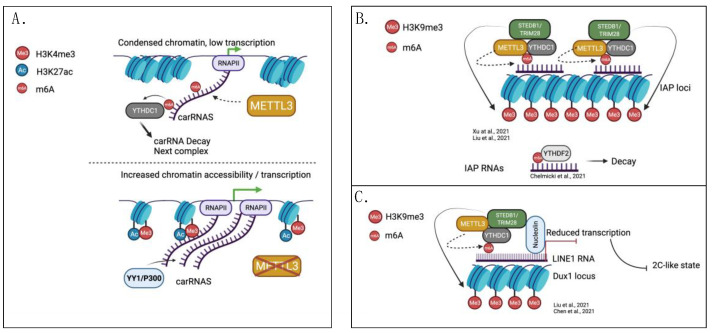
Figure 1.
The regulation of transposable elements and chromatin accessibility by m6A. (A) METTL3-dependent methylation of carRNAs maintains condensed chromatin at intergenic regions and promotes carRNAs degradation by the NEXT (nuclear exosome targeting) complex. Loss of carRNA methylation leads to increased chromatin accessibility and enriched transcription, associated with increased histone H3-lysine4 trimethylation (H3K4me3) and histone H3-lysine27 acetylation (H3K27ac). carRNAs can now recruit epigenetic factors such as YY1 and EP300 to maintain an open chromatin conformation and downstream transcription. (B) METTL3 deposits m6A on intracisternal A particle (IAP) RNAs. YTHDC1 recognises and binds to methylated IAPs, and in conjunction with METTL3 recruits the histone methyltrasferase SETDB1 and its co-factor TRIM28. This complex establishes histone H3-lysine9 trimethylation and maintains a closed chromatin conformation at IAP loci. This leads to an overall reduction in the transcription of IAP RNAs. m6A-marked IAP RNAs are degraded by YTHDF2. (C) m6A-marked LINE1 silences the Dux1 locus in mouse embryonic stem cells to prevent the activation of the 2C-like state transcriptional program. Methylated LINE1 recruits the methyltransferse SETDB1 and its co-factor TRIM28 through YTHDC1. Nucleolin also takes part in the silencing complex assembled over the m6A-marked LINE1. Created with BioRender.com.