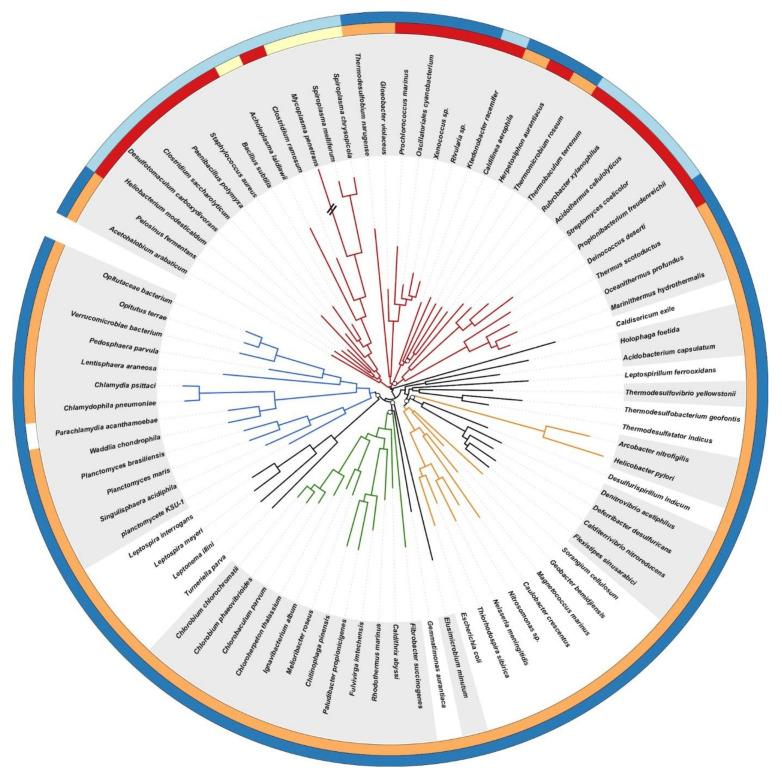
Figure 1.
Phylogenomic tree of the bacterial domain based on a supermatrix concatenating 117 single-copy orthologous genes chosen for their broad conservation across Bacteria. The tree was rooted on Terrabacteria. The supermatrix had 85 species and 19,959 unambiguously aligned amino-acid positions (<5% missing character states). The tree was inferred from amino-acid sequences using PhyloBayes MPI and the CAT+GTR+Γ model of sequence evolution. Open symbols at the nodes are posterior probabilities (PP), and nodes without a symbol correspond to maximum statistical support for phylogenetic inference (posterior probabilities of 1.0; averaged over two MCMC chains). The length of the branch marked with “//” has been reduced by 50% for the sake of clarity. Colour key is red = Terrabacteria, orange = Proteobacteria, green = FBC superphylum, blue = PVC superphylum. Outer circles stand for the status of the peptidoglycan (PG) and of the outer membrane in the organisms, according to our literature survey: red = thick PG, orange = thin PG, yellow = no PG, dark blue = diderm, light blue = monoderm, white = no information. Alternating white and grey backgrounds highlight the alternance between differentially coloured groups or phyla.