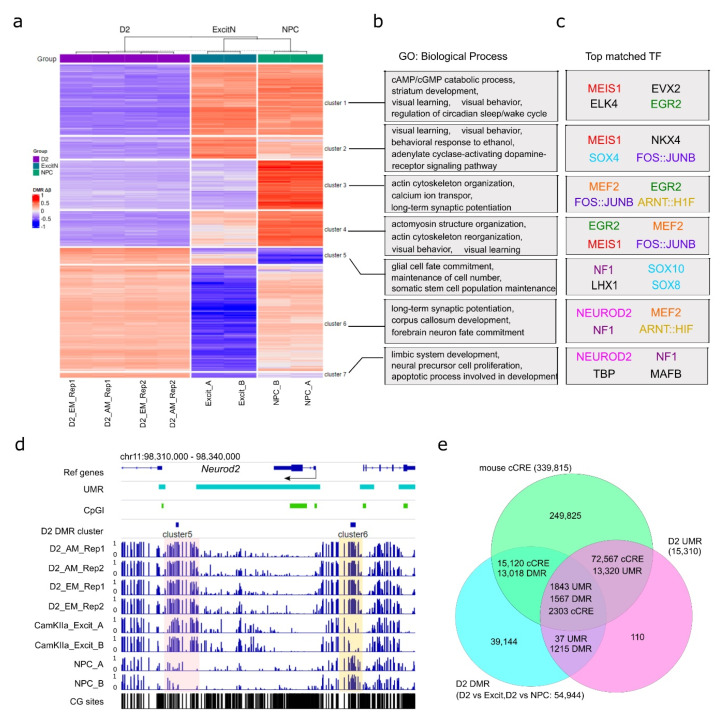
Figure 4.
NAc D2-MSN-specific CpG DMRs. (a) K-means clustering of 51,704 NAc D2-MSN-specific autosomal CpG DMRs, in comparison to NPCs and PFC CamKIIa+ excitatory neurons. Two sets of DMRs are combined. For each DMR, the delta β values of individual samples (compared to the average methylation level across all samples) are used for clustering. (b) Representative biological process gene ontology (GO) terms enriched with genes associated with each cluster of CpG DMRs in panel a. (c) Transcription factors (TFs) with matched sequences of the top 4 motifs enriched in each of the seven CpG DMR clusters. Identical or similar transcription factors are in the same color, except where black is used for motifs only recognized in a single cluster. (d) The genomic locus of Neurod2 is a representative of transcription factor genes enriched in D2-MSN hyper CpG DMRs with D2-MSN-specific hypermethylation. The shaded regions indicate notable cell type-specific differential methylation when compared to NPCs or PFC excitatory neurons. (e) UMRs and cell type-specific DMRs are largely overlapped with mouse cCREs. D2: D2-MSNs. CamKIIa_Excit: PFC CamKIIa+ excitatory neurons. NPC: neural progenitor cells. AM: AM-seq. EM: EM-seq.