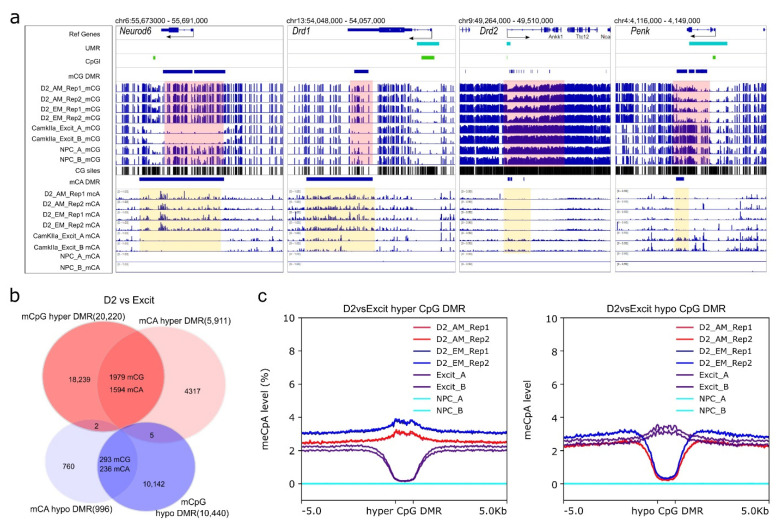
Figure 5.
NAc D2-MSN-specific non-CpG DMRs. All non-CpG DMRs are identified by comparing NAc D2-MSNs to PFC CamKIIa+ excitatory neurons (D2 vs. Excit). (a) Representative view of Drd1 and Neurod6 loci with CpG and CpA hypermethylation in D2-MSNs. In contrast, D2-MSN marker genes Drd2 and Penk have CpG and CpA hypomethylation along their genomic loci when compared to PFC CamKIIa+ excitatory neurons. Pink-shaded regions highlight CpG DMR regions, yellow-shaded regions highlight CpA DMR regions. β value range of all CpG lanes is 0–1. β value range of CpA for Neurod6 and Drd1 is 0–1. β value range of CpA for Drd2 and Penk is 0–0.5. NPC has virtually no CpA methylation. (b) CpG DMRs and CpA DMRs are significantly overlapped. (c) CpA methylation levels at CpG DMRs. Left: CpA methylation levels of D2 hyper-methylated CpG DMRs (blue and red) are noticeably higher than surrounding regions in D2-MSNs and much lower in PFC excitatory neurons (purple). Right: The CpA methylation levels of D2 hypo-methylated CpG DMRs are much lower in D2-MSNs (blue and red) and are discernibly higher in PFC excitatory neurons(purple) compared to the adjacent regions. The NPC methylome is devoid of non-CpG methylation (cyan).