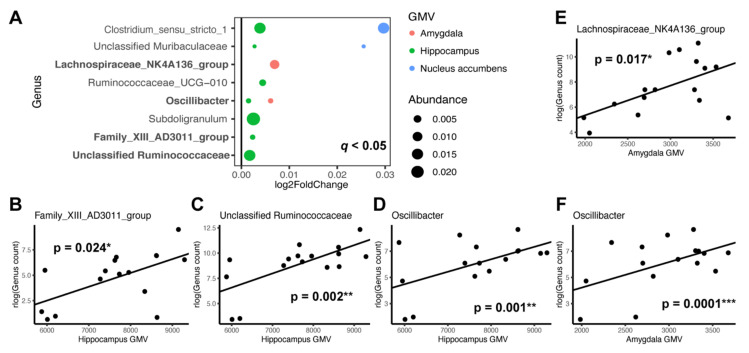
Figure 2.
Specific microbial genera associated with GMV regions. GMV regions tested include hippocampus, amygdala, nucleus accumbens and pericalcarine (control). (A) DESeq2 was used to identify microbial genera associated with GMV with false discovery rate of q < 0.05 after adjusting for age and sex. Log2 fold change represents effect size and direction of these associations. Dot size is proportional to the relative abundance of the genus and color corresponds to the specific brain region. Bolded y-axis text: genera that were confirmed by individual testing of the taxon to be significantly associated with a specific GMV. (B–F) Scatter plots displaying GMV of a specific brain region and regularized log transformed (rlog) normalized count of individual genus with significant associations after adjusting for age and sex (p < 0.05). Regression lines are drawn without controlling for age and sex. * p < 0.05. ** p < 0.01. *** p < 0.001.