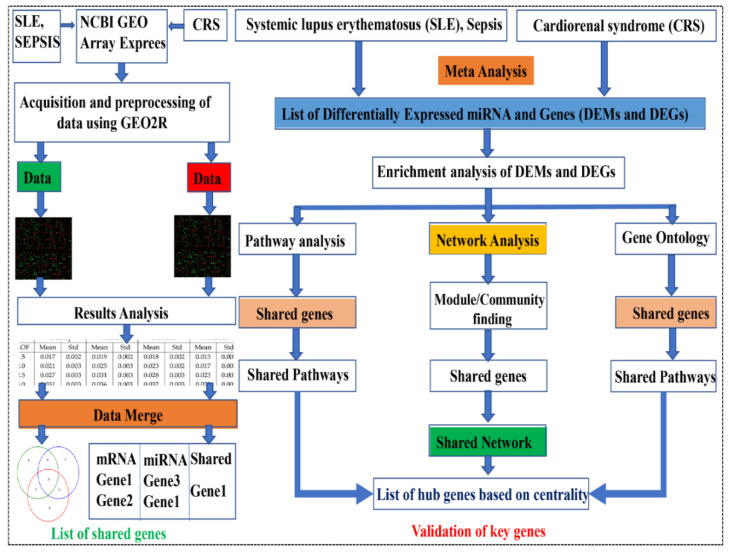
Figure 1.
Methodology adopted for the series used in the study. SLE, sepsis, and CRS data were extracted from NCBI geo datasets, and these terms were also searched for in the Array Express database. The GSE series were available in the public databases, and we retrieved only human data by applying exclusion criteria. Data were filtered via GEO2R and selected after final mining, applying normalization and log 2 transformations. The resulting data are in the Excel sheet used for DEGs (based on fold change and p-value). The data were merged to find overlapping DEGs and DEMs, and finally, the PPIN network was constructed via overlapping DEGs-DEMs (shared network).