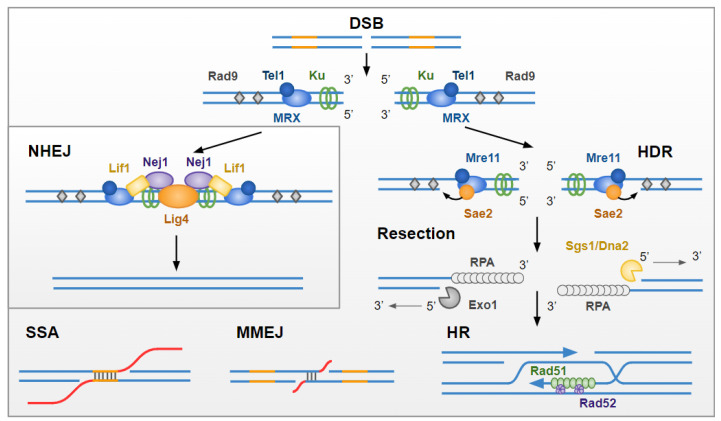
Figure 1.
DNA double-strand break repair pathways. DNA double-strand breaks (DSBs) can be repaired by direct re-ligation of broken ends (non-homologous end joining, NHEJ), or through using a homologous template (homology-directed repair, HDR). On the occurrence of a DSB, DNA damage response factors Ku, MRX, Tel1 and Rad9 are recruited to the damaged site. If repair by NHEJ is favored, Lif1, Nej1 and Lig4 are recruited, and broken DNA is re-ligated (see Table 1 for human orthologs). HDR requires the formation of 3′ single-stranded-DNA (ssDNA) overhangs at the DSB site in a process known as resection. Resection is initiated by the endonuclease activity of Mre11 upon stimulation by Sae2 and proceeds due to the activity of the redundant exonucleases Exo1 and Sgs1/Dna2. The 3′ ssDNA overhangs are stabilized by replication protein A (RPA). Rad52 mediates the replacement of RPA for Rad51. Typically, resected Rad51-bound DSB ends undergo repair by homologous recombination (HR), invading the DNA duplex of the replicated sister chromatid for use as a template for faithful DNA DSB repair. Although NHEJ and HR are the canonical DSB repair pathways, other mechanisms are also observed. Repair by microhomology-mediated end joining (MMEJ) is dependent on short ~4–20 bp homologous sequences situated close to the DSB on either side of the break. These short homologous sequences can anneal with one another, sealing the DSB, but generating small deletions (in red). Alternatively, unmasking of longer direct homologous repeats (in orange) can lead to repair by single-strand annealing (SSA), a process that also sees the loss of the genomic sequence that once separated them (in red).