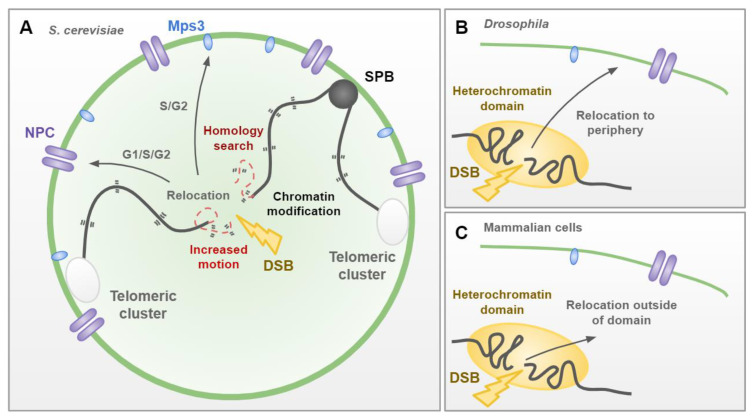
Figure 3.
Chromatin dynamics in response to DNA double-strand breaks (DSBs). (A) In S. cerevisiae, chromosome centromeres are tethered to the spindle pole body (SPB), and telomeres cluster at the nuclear periphery. Upon DSB both local and global processing of the chromatin fiber alter its properties. These chromatin modifications lead to increased chromatin motion of DSB ends and the global genome, which likely assists in the homology search process. Persistent DSBs relocate to the nuclear periphery, through either interaction with the nuclear pore complex (NPC) or Mps3, in a cell-cycle-dependent manner, to assist repair by alternative mechanisms. (B) In Drosophila, heterochromatic DSBs move out of heterochromatin domains and to the nuclear periphery to facilitate faithful repair. (C) In mammalian cells, heterochromatic DSBs move out of heterochromatin domains, but not to the nuclear periphery, to facilitate DSB repair.