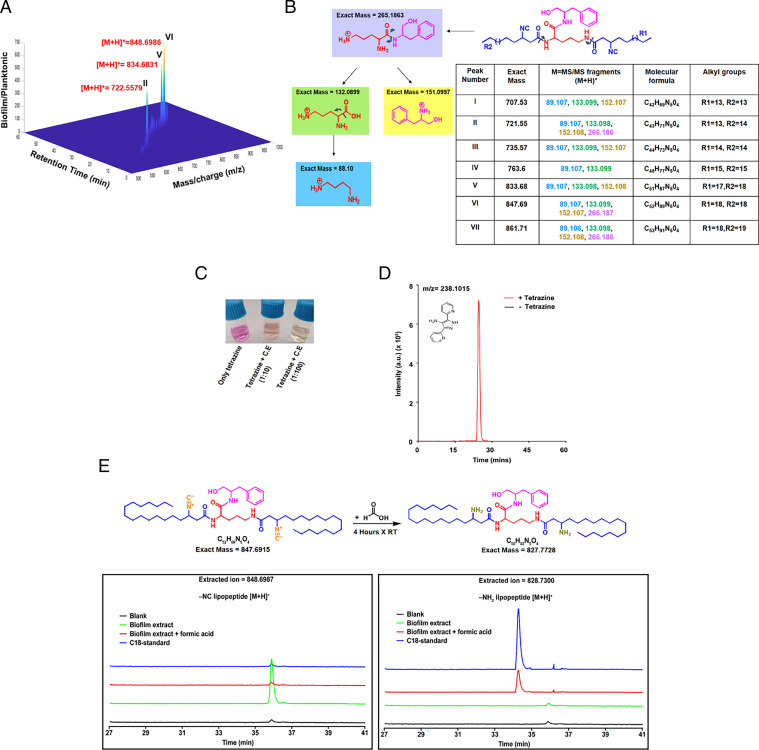
Fig. 5.
Identification and characterization of kupyaphores from Mtb (A) Three-dimensional heat map plot shows masses uniquely identified from biofilm extracts of Mtb cultures but not planktonic cultures at various retention times. Three peaks with the highest fold-change between biofilm and planktonic cultures corresponded to m/z [M+H]+= 722.55 (II), 834.68 (V), and 848.69 (VI), are marked here in red. (B) MS/MS fragmentation pattern revealed common dipeptide backbone of ornithine and phenylalaninol in all the seven kupyaphore species (I–VII) observed in the complete spectra. Common fragments identified in the positive ion mode for all these masses show identical pattern. The predicted fragments are color coded along with their masses in the table. (C) Color change of Py-tetrazine from pink to yellow observed upon addition of WT Mtb biofilm extract. (D) Extracted ion chromatogram (EIC) for [M+H]+ = 238.1014 corresponding to universal product, Py-aminepyrazole, in biofilm culture extract with and without tetrazine addition. (E) MS-based comparative analysis of EIC for biofilm organic extract and synthetic chemical standard at [M+H]+ = 828.7300 and 848.6987. Addition of formic acid to biofilm organic extract shifts the mass of parent ion (m/z-848.6987) to that of synthetic chemical standard of amino substituted lipopeptide (m/z-828.73) concomitant with conversion of isonitrile to amine upon acid hydrolysis. The observed masses for I–VII are within 10 ppm mass error tolerance at MS1 or MS/MS for each mass.