Significance
Flavoproteins are colored proteins involved in a large variety of biochemical reactions. They can perform photochemical reactions, which are increasingly exploited for bioengineering new protein-derived photocatalysts. In particular, light-induced reduction of the resting oxidized state of the flavin by close-lying amino acids or substrates is extensively studied. Here, we demonstrate that the reverse and previously unknown reaction photooxidation of the anionic semireduced flavin radical, a short-lived reaction intermediate in many biochemical reactions, efficiently occurs in flavoprotein oxidases. We anticipate that this finding will allow photoreduction of external reactants and lead to exploration of novel photocatalytic pathways.
Keywords: electron transfer, flavoproteins, ultrafast spectroscopy, molecular modeling
Abstract
The photophysical properties of anionic semireduced flavin radicals are largely unknown despite their importance in numerous biochemical reactions. Here, we studied the photoproducts of these intrinsically unstable species in five different flavoprotein oxidases where they can be stabilized, including the well-characterized glucose oxidase. Using ultrafast absorption and fluorescence spectroscopy, we unexpectedly found that photoexcitation systematically results in the oxidation of protein-bound anionic flavin radicals on a time scale of less than ∼100 fs. The thus generated photoproducts decay back in the remarkably narrow 10- to 20-ps time range. Based on molecular dynamics and quantum mechanics computations, positively charged active-site histidine and arginine residues are proposed to be the electron acceptor candidates. Altogether, we established that, in addition to the commonly known and extensively studied photoreduction of oxidized flavins in flavoproteins, the reverse process (i.e., the photooxidation of anionic flavin radicals) can also occur. We propose that this process may constitute an excited-state deactivation pathway for protein-bound anionic flavin radicals in general. This hitherto undocumented photochemical reaction in flavoproteins further extends the family of flavin photocycles.
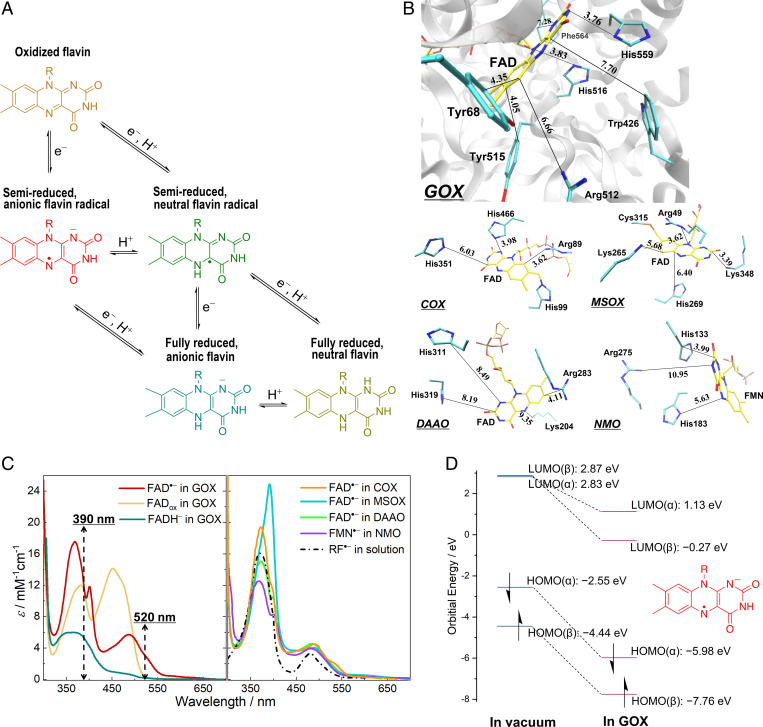
Photoinduced processes involving radicals can have deleterious biological consequences. Yet, radicals play functional roles in biochemical reactions. One group of photochemically active biological macromolecules where radical intermediates can be formed is constituted by flavoproteins. These are ubiquitous components in key biological processes, in particular in redox reactions critical to the major metabolic energy transformations and electron transfer (ET) chains (1–3). The flavin cofactor usually exists as riboflavin (RF), that is, vitamin B2, derivative flavin mononucleotide (FMN), or, more often, flavin adenine dinucleotide (FAD). Flavin groups are versatile redox partners, as they can adopt three different redox states, each with different possible protonation states, overall including five physiologically relevant chemical forms (Fig. 1A) (4).
Fig. 1.
Steady-state properties of protein-bound flavin species. (A) Five redox and protonation states of flavins occurring as physiological reaction intermediates. Flavin is represented by the isoalloxazine moiety. (B) Active sites in the crystal structures of GOX from Aspergillus niger (PDB entry 1CF3), COX from Arthrobacter globiformis (S101A variant; PDB entry 3NNE), MSOX from Bacillus sp. (PDB entry 2GB0), DAAO from porcine kidney (PDB entry 1VE9; in complex with benzoate [not shown]), and NMO from P. aeruginosa PAO1 (PDB entry 4Q4K). The carbon atoms of the flavin cofactors are shown in yellow, whereas those of selected active-site residues are displayed in cyan; nitrogen, oxygen, and sulfur atoms are colored in blue, red, and orange, respectively. The closest distances between flavin rings and nonhydrogen atoms of side chains are shown in angstroms. (C) Steady-state absorption spectra of flavin species in different systems. FAD•− in GOX was generated by photoreduction in the presence of ethylenediaminetetraacetic acid at pH 10.1, and the spectrum of FADox in GOX was measured at pH 10.1. Spectra of FAD•− in DAAO and RF•− in solution were reproduced from published data (26, 42). The other spectra were measured, as described in the SI Appendix, Methods. For all the spectra, the reported values of extinction coefficients were used (26, 27, 30, 31). Dashed lines indicate the maxima of excitation pulses in the time-resolved spectroscopic measurements. (D) HOMO and LUMO energy levels of FAD•− calculated in vacuum and in GOX at the ωB97X-D3/ma-def2-TZVP level.
Flavins are colored molecules, and photochemical processes are known to occur in many flavoproteins; in some specific cases, these processes play a functional role. Photoinduced phenomena of oxidized flavin, the resting state in most flavoproteins, have been studied extensively. Oxidized flavin is strongly fluorescent in solution, but when bound to proteins, its fluorescence is often quenched as it may act as an electron acceptor and participate in photoinduced ET reactions (5–12). Fully reduced (neutral or anionic) flavins emit virtually no fluorescence in solution, but in a confined protein environment, or at low temperature, their fluorescence is greatly enhanced, a phenomenon ascribed to hindering of deactivation by bending motions of the isoalloxazine ring (13, 14). Semireduced (neutral or anionic) flavin radicals are usually short-lived species formed in the catalytic cycles of flavoproteins and in aqueous solution cannot be obtained as free radicals in the steady state. In some flavoproteins where neutral flavin radicals can be stabilized (e.g., DNA photolyase and flavodoxin), excitation has been reported to extract an electron from nearby aromatic residues (15–17); in DNA photolyase, this process generates the long-lived and catalytically active, fully reduced flavin cofactor (18). In contrast, how anionic flavin radicals behave upon excitation remains unclear despite their relevance as functional intermediates in many flavoproteins.
In blue light receptor proteins, such as cryptochromes and blue light using flavin (BLUF) domain proteins, the photoreduction of oxidized flavin (FMNox or FADox) to the anionic radical form (FMN•− or FAD•−) represents the first essential step in their response to light (19, 20). In fatty acid photodecarboxylase, the photoinduced ET between FADox and the fatty acid substrate leads to FAD•− as an intermediate (21). The catalytic activity of the flavoprotein monooxygenase PqsL can be triggered by blue light illumination, which was proposed to occur via photoinduced formation of the NAD(P)H•+/FAD•− radical pair (22). Light-independent processes can also occur through pathways involving anionic flavin radical formation. This is, for instance, the case for the oxidative catalysis of organic molecules by nitronate monooxygenase (NMO) (23) and by glucose oxidase (GOX), which is widely used in biotechnology applications (24, 25). Yet, despite decennia of in-depth flavoprotein research, so far, only a few flavoproteins have been reported to be able to stabilize the anionic flavin radical in the steady state (19, 26–31). As a result, understanding of their photophysical properties is limited.
In general, radical ions in open-shell configurations have distinct photophysical properties from those of their closed-shell parent molecules. Their excited states usually decay in a complex manner, involving efficient internal conversion, or conical intersections between excited state and ground state (32, 33). With electron donors or acceptors close by, ultrafast ET can also take place (34); chemical systems that make use of such an ET reaction from the doublet excited state have been developed as superreductants/superoxidants for solar energy and photocatalytic applications (35–37). The present work provides an ultrafast spectroscopic investigation with full spectral resolution to elucidate the actual deactivation pathway of protein-bound anionic flavin radicals. In particular, we studied a set of different flavoprotein oxidases, where the anionic flavin radicals can be stabilized and of which well-characterized crystal structures are available (i.e., FAD•− in GOX, choline oxidase [COX], monomeric sarcosine oxidase [MSOX], and d-amino acid oxidase [DAAO] as well as FMN•− in NMO. In all these enzymes, the active site contains positively charged residues that are thought to be required for stabilizing the superoxide O2•− functional reaction intermediate (“oxygen activation”) (38, 39). In all systems, these include residues that are close to van der Waals contact with the isoalloxazine ring system (Fig. 1B).
In photoinduced ET processes, flavin species almost always act as electron acceptors and are photoreduced, as is the case with protein-bound oxidized flavins and neutral flavin radicals (5–12, 15–17). The only known exception in nature thus far is DNA photolyase. In the photorepair process of both cyclobutane pyrimidine dimer (CPD) and (6, 4)-photolyases, where the flavin cofactors function in the fully reduced, anionic form FADH–, the repair of ultraviolet (UV)-induced DNA lesions is initiated by photoinduced ET from FADH– to the DNA substrate, yielding the neutral flavin radical (FADH•) (18, 40). Recently, a similar photoreaction in flavin-dependent “ene”-reductases has been exploited as a tool to initialize stereoselective radical cyclization reactions in organic synthesis (41). Our present findings reveal yet another reaction pathway in the photochemistry of flavoproteins, namely, the ultrafast photooxidation of anionic flavin radicals. We establish that excitation of these species results in their photooxidation on a time scale of less than ∼100 fs, generating oxidized flavins in the transient product state that decay in 10 to 20 ps back to the anionic radical forms. As similar phenomena were observed for all five flavoprotein oxidases in our study, such a photooxidation reaction might constitute an intrinsic characteristic of protein-bound anionic flavin radicals in general. We further discuss the identity of potential electron acceptors based on density-functional theory (DFT) calculations and molecular dynamics (MD) simulations as well as hybrid quantum mechanics/molecular mechanics (QM/MM) methods.
Results
Steady-State Absorption Spectra of Anionic Flavin Radicals.
Anionic flavin radicals are usually short-lived reaction intermediates, and in solution, their formation and detection require pulse radiolysis methods (42). In some flavoproteins, anionic flavin radicals can be produced and stabilized under anaerobic conditions by blue light illumination in the presence of a hole scavenger or through chemical reduction (26–28, 30, 31). Fig. 1C shows the absorption spectra of protein-bound anionic flavin radicals produced and stabilized in GOX, COX, MSOX, DAAO, and NMO. In the visible range, anionic flavin radicals exhibit two distinct transition bands (assigned to D0→D4 and D0→D6 transitions) (11). Those obtained in proteins differ significantly from those produced in solution by pulse radiolysis and from each other. This can be explained by the interactions between the anionic flavin radicals and the local protein environments and might be an indication of positive charges close by, as has been demonstrated in our previous work (11). A QM/MM calculation (Fig. 1D) shows that the protein environment of GOX significantly lowers the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) energy levels of FAD•− compared with those in vacuum, indicating the effects of the protein environment on the electronic properties of FAD•−. Furthermore, we note that the presence of small amounts of oxidized or fully reduced flavin in the equilibrated samples cannot be fully excluded (see SI Appendix, Note 1).
Emission from Excited Anionic Flavin Radicals.
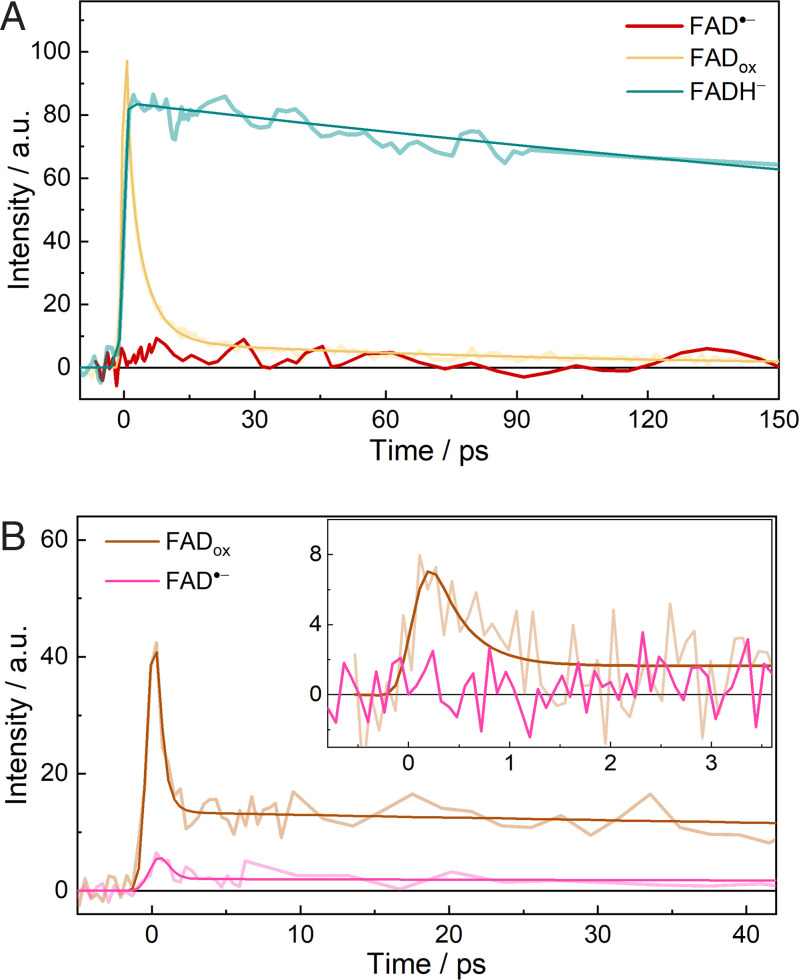
First, we investigated the excited state dynamics of FAD•− using time-resolved fluorescence measurements. As shown in Fig. 2A and SI Appendix, Fig. S5, upon excitation at 390 nm, we did not detect any perceptible emission signal for FAD•− in GOX with our setup that has femtosecond time resolution (for samples obtained either by photo or chemical reduction). The fluorescence decays of FADox and FADH− in GOX measured under the same experimental conditions are presented in the same figure for comparison. The fluorescence of FADox in GOX mainly decays with time constants of 1.1 ps and 4.3 ps (6, 12). The time-resolved fluorescence of FADH− in GOX exhibits monophasic decay with a time constant of 800 ps (SI Appendix, Fig. S6B and Table S2). The absence of an emission signal for FAD•− in GOX indicates that there are nonradiative processes taking place much faster than the instrument response (∼200 fs with Suprasil as Kerr medium).
Fig. 2.
Time-resolved fluorescence measurements. (A) Fluorescence decays of FAD•−, FADox, and FADH− in GOX monitored at 537 nm under the same excitation conditions with CS2 as the Kerr medium. Intensities were normalized based on the absorption of the samples at the excitation wavelength (390 nm). (B) Fluorescence decays of the as-prepared aerobic (FADox) and reduced (FAD•−) samples of COX monitored at 532 nm, with benzene or Suprasil (inset) as Kerr medium. Measurements were conducted with the same sample before and after the chemical reduction. Exponential fits are shown as smooth curves; a.u., arbitrary units.
FAD•− in COX is stable even in the presence of O2 (31), allowing long-duration signal accumulation for detecting small signals. As a reference, the time-resolved fluorescence of nonreduced, FADox-containing (SI Appendix, Fig. S1) COX was measured (Fig. 2B). Upon excitation at 390 nm, it exhibits a biphasic decay with a dominant ∼300-fs ultrafast phase and a long-lived phase (SI Appendix, Fig. S7A). The 300-fs phase was assigned to FADox fluorescence that was quenched by flavin photoreduction processes, with Tyr465 and Trp61 being the potential quenchers (SI Appendix, Fig. S10). The sample was then incubated with Na2S2O4 until the reaction reached maximal FAD•− formation (corresponding to the absorption spectrum shown in Fig. 1C), and further time-resolved fluorescence measurements were performed. With benzene as Kerr medium (relatively high sensitivity but low time resolution), a small signal less than 10% of that of FADox was detected. It has similar kinetic and spectral (maximum at ∼520 nm) features (Fig. 2B and SI Appendix, Fig. S7) to those of FADox and was therefore assigned to a remaining fraction of FADox. However, when Suprasil was used as Kerr medium (low sensitivity but high time resolution), no perceptible emission signal was detected for the reduced sample, whereas the ∼300-fs phase of FADox is clearly resolved in the nonreduced sample (Fig. 2B, Inset). From the comparison of the signals of the reduced and nonreduced samples, we estimate that any FAD•− fluorescence decays in <30 fs. Altogether, we conclude that as in GOX, in COX, ultrafast nonradiative deactivation of excited-state FAD•− occurs.
Transient Formation of Oxidized Flavins upon the Excitation of Anionic Flavin Radicals.
The lack of emission in radical ions is usually explained by efficient internal conversion between D1 and D0 states, involvement of D2/D1 and D1/D0 conical intersections, or photoinduced ET when there is an electron donor or acceptor close by (32–36). To investigate the actual excited-state deactivation pathways of protein-bound anionic flavin radicals, we performed transient absorption measurements. Pump pulses centered at 390 and 520 nm were used to excite the two distinct transition bands.
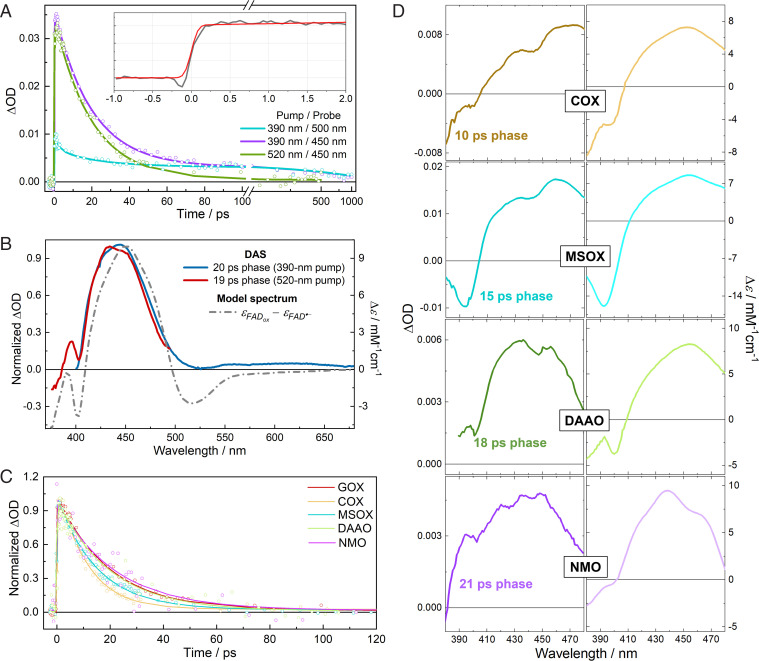
The transient absorption kinetics of FAD•− in GOX are shown in Fig. 3A and SI Appendix, Fig. S13, and the decay associated spectra (DAS) extracted from the global analysis are given in Fig. 3B and SI Appendix, Fig. S15. Here, the DAS were used to avoid the influence of potential parallel photochemical schemes arising from other excitable flavin forms that may be present in the samples (SI Appendix, Note 1). Upon excitation, we observed a marked induced absorption band formed within an instrument-limited time of ∼100 fs (Fig. 3A, Inset). This band is very similar to the S0→S1 transition band of ground-state FADox. Under 520-nm excitation, the decay of this band (time constant of 19 ps) constitutes the only spectral evolution. Under 390-nm excitation, this band has a virtually identical shape (Fig. 3B) and decays with a very similar time constant. Along with the fact that we did not observe any emission with a similar lifetime in the time-resolved fluorescence measurements (vide supra), we conclude that this ∼20-ps phase must arise from a nonemissive product state.
Fig. 3.
Transient absorption measurements. (A) Isotropic transient absorption kinetics of FAD•− in GOX at selected wavelengths upon excitation at 390 nm or 520 nm. Inset, rise of the induced absorption signal (gray) at 433 nm (at the minimum of the probe dispersion curve) under 390-nm excitation. The red curve represents an instrument response–limited rise. Full spectro-temporal data are depicted in SI Appendix, Figs. S13 and S14; ΔOD, change in optical density. (B) Spectral analysis of DAS of FAD•− in GOX under two excitation conditions. The model spectrum was constructed from steady-state spectra by assuming complete FAD•− to FADox conversion. (C) Isotropic transient absorption kinetics in different flavoprotein oxidases after excitation at 520 nm and monitored at 455 nm. (D) DAS of anionic flavin radicals in COX, MSOX, DAAO, and NMO under excitation at 520 nm as well as the corresponding steady-state difference spectra.
The 390-nm excitation data require two additional small kinetic phases with time constants of 1.8 and 900 ps to obtain a satisfactory global fit (Fig. 3A and SI Appendix, Figs. S13 and S15). These time constants are similar to those of the excited-state lifetime of FADox and FADH− (1.1/4.3 ps and 800 ps, respectively, vide supra) in GOX; the corresponding DAS, although too small to be reliably analyzed, also show resemblance to those obtained for FADox (12) and FADH− (SI Appendix, Figs. S11 and S12) (14). Indeed, as illustrated in Fig. 1C, the 390-nm pump will also excite trace amounts of oxidized and fully reduced flavins, whereas the 520-nm pump can only excite anionic flavin radicals.
We observed similar transient absorption spectra for the anionic flavin radicals in the other flavoprotein oxidases. Upon excitation of FAD•− or FMN•− in COX, MSOX, DAAO, and NMO, strong induced absorption bands centered at ∼450 nm appeared in less than ∼100 fs. They highly resemble the S0→S1 transition bands of ground-state FADox or FMNox (Fig. 3 C and D and SI Appendix, Figs. S16 and S17) and upon 520-nm excitation decay in a single-exponential manner in 10 to 20 ps (Fig. 3C and SI Appendix, Table S3). Similar to GOX, experiments with 390-nm excitation on COX and MSOX also yielded minor additional phases assignable to trace amounts of FADox and FADH− (SI Appendix, Note 3).
In Fig. 3 B and D, spectral analysis is performed by comparing the DAS of the product states with steady-state difference spectra of the oxidized flavin and the anionic flavin radical. The overall features of the DAS, including the ground state bleaching and induced absorption, correspond well with the model spectra. Furthermore, taking into account the excitation density using [Ru(bpy)3]Cl2 as a reference, the amplitude of the signals indicates near-unity oxidized flavin formation per absorbed photon (SI Appendix, Note 4).
Yet, the DAS are not strictly identical to the model spectra. For instance, the bleaching feature at ∼500 to 550 nm in the steady-state model spectrum of GOX (Fig. 3B) is less pronounced and nonnegative in the transient spectrum. The discrepancy between the steady-state model and transient spectra may be due to two reasons. First, given the ultrafast time scale, the transiently formed oxidized flavins are presumably in an unrelaxed protein environment, resulting in a shifted absorption spectrum and altered vibrational progression. Second, during the photooxidation process, a radical intermediate formed from a yet unknown electron acceptor may also absorb in the visible region and contribute to the observed DAS. The second assumption (further addressed theoretically in the next section) is supported by the analysis of the transient anisotropy absorption spectra (SI Appendix, Fig. S21), which provides detailed insight in the orientation of the involved transition dipole moments (43) (details for calculating anisotropy r and the angle ϕ between the pumped and probed transition are given in SI Appendix, Note 5). In the 440- to 470-nm spectral range where the transient absorption is maximal, the anisotropies are almost constant and very similar for 390-nm (r = 0.18 to 0.19) and 520-nm (r = 0.20 to 0.22) excitation. As in both cases, mainly the S0→S1 transition band of FADox was probed, this result indicates that the D0→D4 and D0→D6 transition dipole moments of FAD•− in GOX are near parallel, which is in general agreement with the theoretical prediction that these two transition dipole moments form a small angle (SI Appendix, Fig. S22). However, previous work has shown that the S0→S1 transition dipole moment of FADox should also be near parallel to the D0→D4 transition of FAD•−, which corresponds to an r value close to 0.4 (10), whereas smaller values were observed in the present case. This may be explained by broad contributions from transition dipole moments of a third species in the probed region. The identity of this third species is examined in the next section.
Additionally, we investigated whether the photoinduced flavin oxidation in GOX depends on the preparation method of FAD•− or on the pH. No such dependence was observed (SI Appendix, Fig. S19), and this robustness, along with our observations in a multitude of protein systems, strongly suggests that photooxidation of the anionic flavin radical occurs ubiquitously in flavoenzymes where this species can be formed.
Identity of the Electron Acceptor.
With regard to the possible identity of the corresponding electron acceptors, we rule out the involvement of byproducts produced upon preparing the anionic flavin radicals in the steady state, as similar results were obtained with different reduction methods, including photoreduction. The ultrafast formation of the transient oxidized flavins and the monophasic decays suggested a close interaction and a well-defined conformation between the electron donor and acceptor, which makes active-site residues appropriate candidates.
In DFT, the ionization potential (IP) and the electron affinity (EA) of a molecule can be approximated by the negative of the HOMO energy, which, to be more precise, refers to the highest occupied Kohn–Sham (KS) levels here of the N and N + 1 electron systems, respectively (44). For selected active-site residues in the investigated flavoprotein oxidases, DFT calculations were thus carried out to evaluate their EA by optimizing their geometries in the one-electron–reduced forms (N + 1). The result shows that in vacuum, only the neutral radicals of fully protonated histidine (HisH2+) and arginine (ArgH+) have reasonable negative HOMO energies (−5.24 and −5.92 eV, respectively), indicating the relatively high (and close) EA, whereas for the other residues, including the singly protonated forms of histidine, the acceptance of an additional electron leads to relatively unstable species (SI Appendix, Table S4). Indeed, among the one-electron–reduced forms of common amino acids, so far, only the neutral radicals of isolated protonated histidine and arginine (or analogs) have been experimentally produced and spectrally characterized (45, 46), suggesting that their reductions are energetically more accessible. Additionally, several studies have demonstrated that HisH2+ is an effective electron acceptor for quenching excited tryptophan (*Trp) (47, 48). Together, this suggests that HisH2+ and ArgH+ are the most likely candidates for the electron acceptors among those residues. Notably, the neutral lysine radical appears to have a moderately negative HOMO energy (−3.77 eV); the backbone amide group, which has been reported to quench *Trp fluorescence (49) and is present in proximity of the flavins, is determined to have a HOMO energy of −0.60 eV. Therefore, protonated lysine and backbone amide are energetically less favorable to act as an electron acceptor than HisH2+ and ArgH+.
Among the five flavoprotein oxidases, the structural properties of GOX are the most extensively studied and relatively well understood, making it an appropriate system for further theoretical investigation of the electron acceptor. In GOX, there are two histidine residues located very close to the flavin in the active site (Fig. 1B), that is, His516 and His559 (3.83-Å and 3.76-Å ring–to–ring distance to FAD in the crystal structure, respectively). In accord with the DFT calculations above, a doubly protonated form of histidine is more likely to act as electron acceptor; therefore, we examined the protonation state of these two histidines. His559 forms strong hydrogen bonds to Glu412 and is expected to be doubly protonated over a wide pH range (50, 51). However, the protonation state of His516 in the presence of FAD•− is less clear, which was examined by MD free energy (MDFE) simulations. The result (SI Appendix, Table S5) shows that, in the presence of FAD•−, in the doubly protonated form, His516 also has a very high pKa of 13.5 (SD: 0.7) due to interactions with the negatively charged FAD•− and nearby Asp328. Therefore, in GOX, both histidine residues, His516 and His559, are likely to be doubly protonated in the presence of FAD•−. The active site of GOX appears to be arranged in a way that in the reduced form, the pKa of the flavin cofactor is downshifted (52), while the pKa values of the two histidines are upshifted.
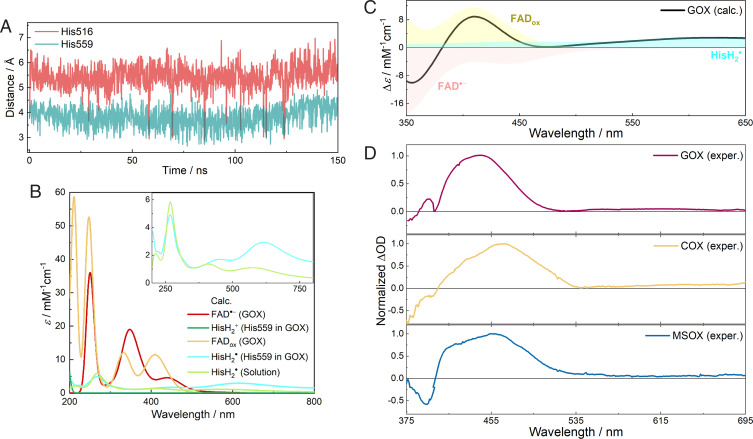
MD simulations were then performed for GOX with the flavin in the anionic radical state and both His516 and His559 doubly protonated. As shown in Fig. 4A, throughout the 150-ns MD simulation, His559 remains very close to FAD•− with an average minimal ring-to-ring distance of 3.81 Å. However, His516 rotates around χ2 to a noncatalytic conformation, as it was shown previously (53), and stays relatively far away from FAD•− (∼5.5 Å) and only transiently approaches the cofactor. The shorter distance of 3.83 Å between flavin and His516 observed in the crystal structure is explained by the fact that His516 is singly protonated with FADox in the crystal structure (50, 53). The finding of His516 conformational changes is in agreement with a previous study showing that His516 is a flexible, functionally important residue that can flip between catalytic and noncatalytic conformations (53). Given the closer interactions of His559 with FAD•−, His559 appears to be a suitable candidate for accepting an electron from excited FAD•−. These results were also reproduced well by the simulations based on a different high-resolution crystal structure (SI Appendix, Fig. S23; Protein Data Bank (PDB) entry 3QVP, resolution of 1.2 Å).
Fig. 4.
MD simulations and QM/MM spectral calculations. (A) The minimal ring-to-ring distance between FAD•− and His516 or His559 in the MD simulation of GOX. (B) Calculated absorption spectra of individual species involved in the photooxidation process of FAD•− in GOX. (C) Calculated transient spectrum of FAD•− photooxidation in GOX by superimposing individual spectra of corresponding components in B. (D) Experimental DAS (10- to 20-ps phases) of FAD•− in GOX, COX, and MSOX obtained by combining normalized DAS from two excitation conditions (375–420 nm: 520-nm pump, 420 to 695 nm: 390-nm pump; see Fig. 3B). Wavelength ranges of calculated and experimental spectra are different to account for a rigid shift arising from the limitation of calculations (11, 78).
Assuming His559 as the electron acceptor, we used a QM/MM protocol to calculate the transient absorption spectra of FAD•− photooxidation in GOX, including the induced absorption of FADox and HisH2• as well as the ground-state bleaching of FAD•− (∼500 to 550 nm; HisH2+ only absorbs in the UV range). As shown in Fig. 4 B and C, despite some overestimations of the transition energies, the calculated spectra of FADox and FAD•− agree well with the experimental spectra (Fig. 1C). In the near UV range, the calculated spectrum of HisH2• in aqueous solution agrees well with the experimental spectrum obtained by pulse radiolysis (45, 54). Notably, in GOX, HisH2• formed from His559 displays more intense absorption in the visible/near-infrared range than its state in solution (Fig. 4B, Inset) presumably due to effects of the complex protein environment around His559, in particular the hydrogen-bonded Glu412. This partially compensates the bleaching of FAD•− and leads to a red-extending tail in the overall difference spectrum, which reproduces the experimental DAS reasonably well (Fig. 4D).
The free energy change associated with the FAD•−/HisH2+ → FADox/HisH2• ET reaction in GOX was estimated using the following method. In a protein system, the free energy change of the investigated ET process (ΔGET,prot) can be calculated by evaluating the electrostatic free energy difference of the two states relative to a reference system in the aqueous solvent:
where ΔGET,solv is the free energy change of the investigated ET process in the aqueous solution obtained by high-level QM calculations (55, 56), and ΔΔGES is the double difference of electrostatic energies of the systems before (FAD•−/HisH2+) and after (FADox/HisH2•) the ET reaction, respectively, evaluated using MD simulations and the Poisson–Boltzmann (PB) method (57) (SI Appendix, Note 7) in the protein relative to the aqueous solvent. Based on this approach, we obtained a ΔGET,prot value of 2.19 eV (SD: 0.05), which is less than the energy of a 520-nm photon (2.38 eV, corresponding to the red edge of the FAD•− absorption), confirming that such a photoinduced ET reaction is energetically feasible. It is also worth noting that the protein environment is found to energetically disfavor the formation of the FADox/HisH2• pair with ΔΔGES = +0.23 eV (SD: 0.05), hinting at the stabilization of the charges of FAD•− and His559+ by surrounding residues in the active site of GOX.
Discussion
Taken together, our time-resolved fluorescence and transient absorption measurements provide a consistent, yet unexpected, picture that ultrafast photooxidation of anionic flavin radicals occurs in GOX, COX, MSOX, DAAO, and NMO. In all systems this process is followed by back transfer of the ejected electron on the picosecond timescale, as evidenced from the monophasic decay of the induced absorption band (Fig. 3C) and the absence of bleaching signals of the anionic flavin radical on longer timescales (Fig. 3A). The very similar observations among the five different oxidases indicate that the ET reactions involve similar distances and electronic properties. As illustrated in Fig. 1B and summarized in SI Appendix, Table S3, in the five flavoprotein oxidases investigated in this study (and possibly for all other functional flavoprotein oxidases), there is always a positively charged residue, including either histidine or arginine, present in the active site (23, 38, 58, 59). Our present results suggest that these residues not only play an important role in the thermodynamic stabilization of anionic flavin radicals (39) but also contribute to the deactivation of the excited-state anionic flavin radicals via a charge-transfer (CT) state. As the presence of close-lying positively charged residues can be considered a prerequisite for the formation and stabilization of anionic flavin radicals in proteins, the photooxidation process may constitute an excited-state deactivation pathway for protein-bound anionic flavin radicals in general. Qualitatively, such properties may also be related to the common requirements of these oxidases to stabilize the electron-rich intermediates in their catalytic cycles (38), including the highly reactive reaction intermediate O2•− in the active site, by a positive charge in close proximity to the flavin (39, 51).
In none of the investigated systems have we been able to detect fluorescence assignable to the anionic flavin radicals, in agreement with the extremely rapid appearance of the oxidized flavin photoproduct. For COX, our assessment of lack of time-resolved FAD•− fluorescence contrasts with the report of FAD•−-assigned steady-state fluorescence peaking around 450 nm (at the blue of the 500-nm FAD•− absorption maximum, that is, violating the Kasha–Vavilov rule) in the enzyme from Arthrobacter globiformis (31). We note that, unfortunately, a fluorescence excitation spectrum was not provided in ref. 31, and C4a-flavin adducts, which can form in COX (60), strongly fluoresce in this region (61, 62). The only time-resolved fluorescence assigned to the anionic flavin radical remains that from insect cryptochrome (28). Yet, the spectral characteristics are very close to those of oxidized flavin, as discussed in ref. 27. Indeed, the reported absorption spectrum of FAD•− in this protein (28) is significantly different from another report of FAD•− in Drosophila cryptochrome (19) in that it shows an unusual plateau at ∼450 nm, near the absorption maximum of FADox (SI Appendix, Fig. S4).
The lifetimes of the formed photoproducts in the flavoprotein oxidases studied span a remarkably narrow range of ∼10 to 20 ps, with three of the lifetimes being at 20 ps within experimental error. This finding may be linked to the very similar and close distances between the flavins and nearby histidine or arginine residues in the five flavoprotein oxidases (Fig. 1B) as well as the similar EA of histidine and arginine residues (SI Appendix, Table S4). In this context, we note that in COX, the covalently linked and functionally relevant histidine His99 (63) is most probably neutral, since in the crystal structure, its Nδ atom does not appear to have any hydrogen bond partners, and the pKa of 8α-N-imidazole–substituted flavins is determined to be ∼6 (N of the imidazole moiety) in solution (64). Therefore, it is not a likely electron acceptor, making His466 a more probable candidate.
HisH2+ can quench Trp fluorescence by accepting an electron from *Trp (47). In the villin headpiece, this reaction occurs between close-lying solvent-exposed residues on a much slower (∼500-ps) timescale (48) than the intraprotein *FAD•− quenching reaction characterized here. The energetics involved are difficult to compare directly (we note that in the gas phase, *FAD•− should be a much stronger reductant than *Trp; the IP of indole is 7.76 eV (65), while the EA of oxidized flavin is 1.86 eV (66), not compensating for the lower energy difference between the excited-state levels [1.75 eV]; these potentials are very different and environment sensitive in solution and protein). Additionally, and importantly, one may expect that compared with the fluctuating Trp/HisH2+ system in the villin headpiece, the FAD•−/HisH2+ system has a much stronger electronic coupling due to a much more confined environment.
Finally, we emphasize that we did not directly observe the formation of the excited state of anionic flavin radicals (Fig. 3), leaving room for the discussion of the involved ET mechanism. Although our present simulations demonstrate that such a reaction should be energetically feasible, given the estimated weak driving force (∼0.2 eV), it would be incompatible with a standard Marcus theory description using a reorganization energy in the 0.7- to 1-eV range (67, 68). However, on this timescale and with strong ionic interactions between the reactants, the underlying assumptions of thermalized reaction-coordinate motions and weak coupling are likely not fulfilled. Furthermore, we cannot completely rule out a CT excitation that leads to direct formation of a FADox/HisH2• CT state nor nonadiabatic transition from the locally excited state to a CT state. Nevertheless, a direct CT transition, which may overlap with the local excitation transition (compare with ref. 69), in principle requires very strong interactions between the donor–acceptor pair in the ground state to provide reasonable oscillator strength, which is difficult to evaluate using the present level of simulations. It is of note that the crystal structures of the proteins studied a priori correspond to those with oxidized flavins and additionally for COX to a protein variant and for DAAO to a protein–inhibitor complex. When the flavins are reduced, the active-site conformations are likely to contain rearrangements. This makes quantitative evaluation of ET dynamics and a definitive determination of the electron acceptor(s) challenging at this point. Moreover, variants of the close-lying positively charged residues are likely to influence the formation and stabilization of anionic flavin radicals as well as affect the binding affinity for flavins (51, 59, 70). Future studies combining more extensive theoretical investigations and spectroscopic experiments on genetically modified proteins will allow determining the exact nature of the electron acceptors and ultimately establish the precise molecular origin of the common dynamic features of ET in flavoprotein oxidases.
In summary, in this work, we have demonstrated a photochemical reaction occurring in the anionic radical state of flavoprotein oxidases, further extending the variety of light-induced processes in this important class of enzymes (Fig. 5). In view of the ephemeral nature of anionic flavin radicals in most biochemical and photobiochemical reactions where it could act as an intermediate, whether the here-characterized photoreaction could play a physiological or photoprotective role in native proteins remains to be established. Yet, it is plausible that anionic flavin radicals can be accumulated under reducing and low-oxygen physiological conditions. By genetic engineering approaches, we envisage to explore the possibilities of creating pathways that allow further electron hopping, thus stabilizing the primary photoproduct. These approaches ultimately will allow photoreduction of external reactants and pave the way toward the development of a novel class of flavoprotein photocatalysts.
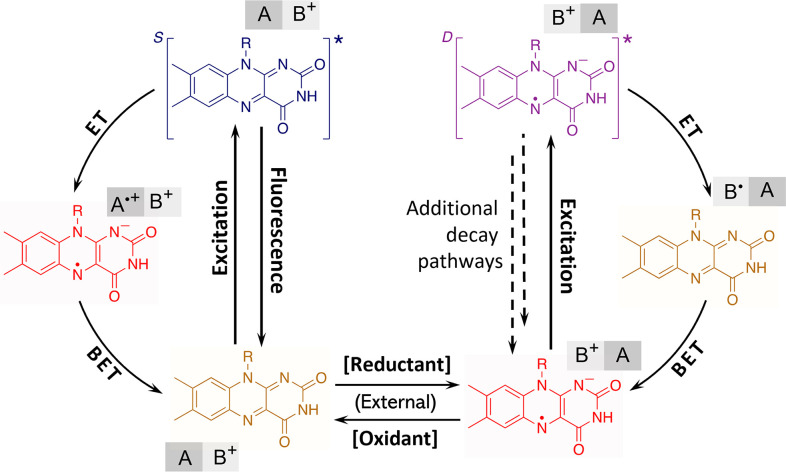
Fig. 5.
General reaction pathways for light-induced processes in flavoproteins that include the proposed cycle of the photooxidation of anionic flavin radicals. Formation of the doublet excited state and potential additional decay pathways (dashed lines) were not directly observed in this study. Flavin is represented by the isoalloxazine moiety. A stands for the electron-donating residue (tryptophan or tyrosine,) and B+ represents the electron-accepting residue (presumably histidine or arginine) in the protein active site; BET: back ET.
Materials and Methods
GOX from Aspergillus niger, COX from Arthrobacter sp., MSOX from Bacillus sp., and DAAO from porcine kidney were purchased from Sigma-Aldrich. NMO from Pseudomonas aeruginosa was expressed using a codon-optimized expression vector and purified following standard procedures. The anionic flavin radicals in these oxidases were prepared by previously reported methods with slight modifications (26, 27, 71).
The setup for time-resolved and spectrally resolved fluorescence uses a Kerr gate (72), and multicolor time-resolved absorption spectra were recorded by the pump–probe technique (9, 10) based on an instrument operating at 500 Hz. Global analysis of the data was performed using the Glotaran program (73).
DFT calculations were carried out using the ORCA program (version 4.2) (74) to optimize the geometries and estimate the energies of studied molecules. For GOX, MDFE simulations for calculating pKa values and full-protein MD simulations were carried out using the NAMD program (version 2.13) (75). Hybrid QM/MM calculations were performed using the pDynamo program (version 1.9.0) (76) coupled with the ORCA package to obtain the KS levels and further estimate the spectral properties of studied molecules based on time-dependent DFT. The PB equation was solved using the CHARMM program (version 45b2) (77) to evaluate electrostatic energies in the protein and solution for computing free energy changes associated with redox processes.
A detailed description of the experimental procedures, data analysis, and simulation protocols is given in the SI Appendix, Methods.
Supplementary Material
Acknowledgments
B.Z. thanks the China Scholarship Council for providing a PhD scholarship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.T. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118924119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Walsh C., Flavin coenzymes: At the crossroads of biological redox chemistry. Acc. Chem. Res. 13, 148–155 (1980). [Google Scholar]
- 2.Fraaije M. W., Mattevi A., Flavoenzymes: Diverse catalysts with recurrent features. Trends Biochem. Sci. 25, 126–132 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Dijkman W. P., de Gonzalo G., Mattevi A., Fraaije M. W., Flavoprotein oxidases: Classification and applications. Appl. Microbiol. Biotechnol. 97, 5177–5188 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Miura R., Versatility and specificity in flavoenzymes: Control mechanisms of flavin reactivity. Chem. Rec. 1, 183–194 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Mataga N., Chosrowjan H., Shibata Y., Tanaka F., Ultrafast fluorescence quenching dynamics of flavin chromophores in protein nanospace. J. Phys. Chem. B 102, 7081–7084 (1998). [Google Scholar]
- 6.Zhong D., Zewail A. H., Femtosecond dynamics of flavoproteins: Charge separation and recombination in riboflavine (vitamin B2)-binding protein and in glucose oxidase enzyme. Proc. Natl. Acad. Sci. U.S.A. 98, 11867–11872 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nunthaboot N., et al. , Simultaneous analysis of ultrafast fluorescence decays of FMN binding protein and its mutated proteins by molecular dynamic simulation and electron transfer theory. J. Phys. Chem. B 112, 13121–13127 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Laptenok S. P., et al. , Ultrafast real-time visualization of active site flexibility of flavoenzyme thymidylate synthase ThyX. Proc. Natl. Acad. Sci. U.S.A. 110, 8924–8929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacombat F., et al. , Ultrafast oxidation of a tyrosine by proton-coupled electron transfer promotes light activation of an animal-like cryptochrome. J. Am. Chem. Soc. 141, 13394–13409 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Nag L., Sournia P., Myllykallio H., Liebl U., Vos M. H., Identification of the TyrOH•+ radical cation in the flavoenzyme TrmFO. J. Am. Chem. Soc. 139, 11500–11505 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Zhuang B., Seo D., Aleksandrov A., Vos M. H., Characterization of light-induced, short-lived interacting radicals in the active site of flavoprotein ferredoxin-NADP+ oxidoreductase. J. Am. Chem. Soc. 143, 2757–2768 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Nag L., Lukacs A., Vos M. H., Short-lived radical intermediates in the photochemistry of glucose oxidase. ChemPhysChem 20, 1793–1798 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Visser A. J., Ghisla S., Massey V., Müller F., Veeger C., Fluorescence properties of reduced flavins and flavoproteins. Eur. J. Biochem. 101, 13–21 (1979). [DOI] [PubMed] [Google Scholar]
- 14.Enescu M., Lindqvist L., Soep B., Excited-state dynamics of fully reduced flavins and flavoenzymes studied at subpicosecond time resolution. Photochem. Photobiol. 68, 150–156 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Pan J., et al. , Excited-state properties of flavin radicals in flavoproteins: Femtosecond spectroscopy of DNA photolyase, glucose oxidase, and flavodoxin. J. Phys. Chem. B 108, 10160–10167 (2004). [Google Scholar]
- 16.Kundu M., He T. F., Lu Y., Wang L., Zhong D., Short-range electron transfer in reduced flavodoxin: Ultrafast nonequilibrium dynamics coupled with protein fluctuations. J. Phys. Chem. Lett. 9, 2782–2790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukacs A., Eker A. P. M., Byrdin M., Brettel K., Vos M. H., Electron hopping through the 15 A triple tryptophan molecular wire in DNA photolyase occurs within 30 ps. J. Am. Chem. Soc. 130, 14394–14395 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Brettel K., Byrdin M., Reaction mechanisms of DNA photolyase. Curr. Opin. Struct. Biol. 20, 693–701 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Berndt A., et al. , A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 282, 13011–13021 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Gauden M., et al. , Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10895–10900 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorigué D., et al. , Mechanism and dynamics of fatty acid photodecarboxylase. Science 372, eabd5687 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Ernst S., Rovida S., Mattevi A., Fetzner S., Drees S. L., Photoinduced monooxygenation involving NAD(P)H-FAD sequential single-electron transfer. Nat. Commun. 11, 2600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gadda G., Francis K., Nitronate monooxygenase, a model for anionic flavin semiquinone intermediates in oxidative catalysis. Arch. Biochem. Biophys. 493, 53–61 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Porter D. J., Bright H. J., Mechanism of oxidation of nitroethane by glucose oxidase. J. Biol. Chem. 252, 4361–4370 (1977). [PubMed] [Google Scholar]
- 25.Bankar S. B., Bule M. V., Singhal R. S., Ananthanarayan L., Glucose oxidase—An overview. Biotechnol. Adv. 27, 489–501 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Massey V., Palmer G., On the existence of spectrally distinct classes of flavoprotein semiquinones. A new method for the quantitative production of flavoprotein semiquinones. Biochemistry 5, 3181–3189 (1966). [DOI] [PubMed] [Google Scholar]
- 27.Su D., Kabir M. P., Orozco-Gonzalez Y., Gozem S., Gadda G., Fluorescence properties of flavin semiquinone radicals in nitronate monooxygenase. ChemBioChem 20, 1646–1652 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Kao Y.-T., et al. , Ultrafast dynamics and anionic active states of the flavin cofactor in cryptochrome and photolyase. J. Am. Chem. Soc. 130, 7695–7701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina M., Vrielink A., Cammack R., ESR and electron nuclear double resonance characterization of the cholesterol oxidase from Brevibacterium sterolicum in its semiquinone state. Eur. J. Biochem. 222, 941–947 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Wagner M. A., Trickey P., Chen Z. W., Mathews F. S., Jorns M. S., Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants. Biochemistry 39, 8813–8824 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Ghanem M., Fan F., Francis K., Gadda G., Spectroscopic and kinetic properties of recombinant choline oxidase from Arthrobacter globiformis. Biochemistry 42, 15179–15188 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Grilj J., Laricheva E. N., Olivucci M., Vauthey E., Fluorescence of radical ions in liquid solution: Wurster’s blue as a case study. Angew. Chem. Int. Ed. Engl. 50, 4496–4498 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Breslin D. T., Fox M. A., Excited-state behavior of thermally stable radical ions. J. Phys. Chem. 98, 408–411 (1994). [Google Scholar]
- 34.Fujitsuka M., Kim S. S., Lu C., Tojo S., Majima T., Intermolecular and intramolecular electron transfer processes from excited naphthalene diimide radical anions. J. Phys. Chem. B 119, 7275–7282 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Christensen J. A., et al. , Phenothiazine radical cation excited states as super-oxidants for energy-demanding reactions. J. Am. Chem. Soc. 140, 5290–5299 (2018). [DOI] [PubMed] [Google Scholar]
- 36.La Porte N. T., et al. , Photoexcited radical anion super-reductants for solar fuels catalysis. Coord. Chem. Rev. 361, 98–119 (2018). [Google Scholar]
- 37.MacKenzie I. A., et al. , Discovery and characterization of an acridine radical photoreductant. Nature 580, 76–80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghanem M., Gadda G., Effects of reversing the protein positive charge in the proximity of the flavin N(1) locus of choline oxidase. Biochemistry 45, 3437–3447 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Gadda G., Oxygen activation in flavoprotein oxidases: The importance of being positive. Biochemistry 51, 2662–2669 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Domratcheva T., Neutral histidine and photoinduced electron transfer in DNA photolyases. J. Am. Chem. Soc. 133, 18172–18182 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Biegasiewicz K. F., et al. , Photoexcitation of flavoenzymes enables a stereoselective radical cyclization. Science 364, 1166–1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Land E. J., Swallow A. J., One-electron reactions in biochemical systems as studied by pulse radiolysis. II. Riboflavin. Biochemistry 8, 2117–2125 (1969). [DOI] [PubMed] [Google Scholar]
- 43.Parson W. W., Modern Optical Spectroscopy: With Exercises and Examples From Biophysics and Biochemistry (Springer, Berlin, Germany, ed. 2, 2015). [Google Scholar]
- 44.Tsuneda T., Song J. W., Suzuki S., Hirao K., On Koopmans’ theorem in density functional theory. J. Chem. Phys. 133, 174101 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Rao P. S., Simio M., Hayon E., Slmic M., Pulse radiolysis study of imidazole and histidine in water. J. Phys. Chem. 79, 1260–1263 (1975). [Google Scholar]
- 46.Hao C., Seymour J. L., Tureček F., Electron super-rich radicals in the gas phase. A neutralization-reionization mass spectrometric and ab initio/RRKM study of diaminohydroxymethyl and triaminomethyl radicals. J. Phys. Chem. A 111, 8829–8843 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Chen Y., Barkley M. D., Toward understanding tryptophan fluorescence in proteins. Biochemistry 37, 9976–9982 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Tusell J. R., Callis P. R., Simulations of tryptophan fluorescence dynamics during folding of the villin headpiece. J. Phys. Chem. B 116, 2586–2594 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Parson W. W., Competition between tryptophan fluorescence and electron transfer during unfolding of the villin headpiece. Biochemistry 53, 4503–4509 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Wohlfahrt G., et al. , 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes. Acta Crystallogr. D Biol. Crystallogr. 55, 969–977 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Roth J. P., Klinman J. P., Catalysis of electron transfer during activation of O2 by the flavoprotein glucose oxidase. Proc. Natl. Acad. Sci. U.S.A. 100, 62–67 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanner C., Macheroux P., Rüterjans H., Müller F., Bacher A., 15N- and 13C-NMR investigations of glucose oxidase from Aspergillus niger. Eur. J. Biochem. 196, 663–672 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Petrović D., Frank D., Kamerlin S. C. L., Hoffmann K., Strodel B., Shuffling active site substate populations affects catalytic activity: The case of glucose oxidase. ACS Catal. 7, 6188–6197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faraggi M., Tal Y., The reaction of the hydrated electron with amino acids, peptides, and proteins in aqueous solution. II. Formation of radicals and electron transfer reactions. Radiat. Res. 62, 347–356 (1975). [PubMed] [Google Scholar]
- 55.Namazian M., Coote M. L., Accurate calculation of absolute one-electron redox potentials of some para-quinone derivatives in acetonitrile. J. Phys. Chem. A 111, 7227–7232 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Neugebauer H., Bohle F., Bursch M., Hansen A., Grimme S., Benchmark study of electrochemical redox potentials calculated with semiempirical and DFT methods. J. Phys. Chem. A 124, 7166–7176 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Ishikita H., Knapp E. W., Redox potential of quinones in both electron transfer branches of photosystem I. J. Biol. Chem. 278, 52002–52011 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Mattevi A., et al. , Crystal structure of d-amino acid oxidase: A case of active site mirror-image convergent evolution with flavocytochrome b2. Proc. Natl. Acad. Sci. U.S.A. 93, 7496–7501 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghanem M., Gadda G., On the catalytic role of the conserved active site residue His466 of choline oxidase. Biochemistry 44, 893–904 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Orville A. M., Lountos G. T., Finnegan S., Gadda G., Prabhakar R., Crystallographic, spectroscopic, and computational analysis of a flavin C4a-oxygen adduct in choline oxidase. Biochemistry 48, 720–728 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamdane D., Guérineau V., Un S., Golinelli-Pimpaneau B., A catalytic intermediate and several flavin redox states stabilized by folate-dependent tRNA methyltransferase from Bacillus subtilis. Biochemistry 50, 5208–5219 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Sournia P., “La méthylation flavine-dépendante d’acides nucléiques: aspects évolutifs, métaboliques, biochimiques et spectroscopiques,” PhD thesis, École Polytechnique, Palaiseau, France: (2016). [Google Scholar]
- 63.Quaye O., Cowins S., Gadda G., Contribution of flavin covalent linkage with histidine 99 to the reaction catalyzed by choline oxidase. J. Biol. Chem. 284, 16990–16997 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williamson G., Edmondson D. E., Effect of pH on oxidation-reduction potentials of 8 alpha-N-imidazole-substituted flavins. Biochemistry 24, 7790–7797 (1985). [DOI] [PubMed] [Google Scholar]
- 65.Hager J. W., Wallace S. C., Two-laser photoionization supersonic jet mass spectrometry of aromatic molecules. Anal. Chem. 60, 5–10 (2002). [Google Scholar]
- 66.Zhang T., Papson K., Ochran R., Ridge D. P., Stability of flavin semiquinones in the gas phase: The electron affinity, proton affinity, and hydrogen atom affinity of lumiflavin. J. Phys. Chem. A 117, 11136–11141 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Marcus R. A., Sutin N., Electron transfers in chemistry and biology. Biochim. Biophys. Acta Rev. Bioenerg. 811, 265–322 (1985). [Google Scholar]
- 68.Page C. C., Moser C. C., Chen X., Dutton P. L., Natural engineering principles of electron tunnelling in biological oxidation–reduction. Nature 402, 47–52 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Sen T., et al. , Interplay between locally excited and charge transfer states governs the photoswitching mechanism in the fluorescent protein Dreiklang. J. Phys. Chem. B 125, 757–770 (2021). [DOI] [PubMed] [Google Scholar]
- 70.Hassan-Abdallah A., Zhao G., Chen Z. W., Mathews F. S., Schuman Jorns M., Arginine 49 is a bifunctional residue important in catalysis and biosynthesis of monomeric sarcosine oxidase: A context-sensitive model for the electrostatic impact of arginine to lysine mutations. Biochemistry 47, 2913–2922 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Hense A., Herman E., Oldemeyer S., Kottke T., Proton transfer to flavin stabilizes the signaling state of the blue light receptor plant cryptochrome. J. Biol. Chem. 290, 1743–1751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laptenok S. P., Nuernberger P., Lukacs A., Vos M. H., “Subpicosecond Kerr–Gate Spectrofluorometry” in Methods in Molecular Biology, Fluorescence Spectroscopy and Microscopy: Methods and Protocols, Engelborghs Y., Visser A. J. W. G., Eds. (Humana, New York, NY, 2014), vol. 1076, pp. 321–336. [DOI] [PubMed] [Google Scholar]
- 73.Snellenburg J. J., Laptenok S. P., Seger R., Mullen K. M., van Stokkum I. H. M., Glotaran: A Java-based graphical user interface for the R Package TIMP. J. Stat. Softw. 49, 1–22 (2012). [Google Scholar]
- 74.Neese F., The ORCA program system. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2, 73–78 (2012). [Google Scholar]
- 75.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Field M. J., The pDynamo program for molecular simulations using hybrid quantum chemical and molecular mechanical potentials. J. Chem. Theory Comput. 4, 1151–1161 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Brooks B. R., et al. , CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983). [Google Scholar]
- 78.Fabian J., TDDFT-calculations of Vis/NIR absorbing compounds. Dyes Pigm. 84, 36–53 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.