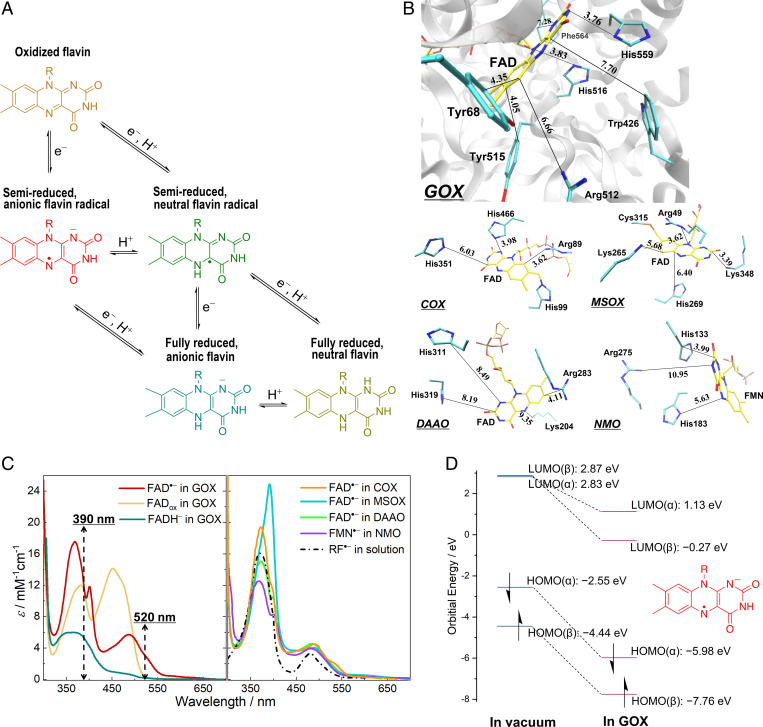
Fig. 1.
Steady-state properties of protein-bound flavin species. (A) Five redox and protonation states of flavins occurring as physiological reaction intermediates. Flavin is represented by the isoalloxazine moiety. (B) Active sites in the crystal structures of GOX from Aspergillus niger (PDB entry 1CF3), COX from Arthrobacter globiformis (S101A variant; PDB entry 3NNE), MSOX from Bacillus sp. (PDB entry 2GB0), DAAO from porcine kidney (PDB entry 1VE9; in complex with benzoate [not shown]), and NMO from P. aeruginosa PAO1 (PDB entry 4Q4K). The carbon atoms of the flavin cofactors are shown in yellow, whereas those of selected active-site residues are displayed in cyan; nitrogen, oxygen, and sulfur atoms are colored in blue, red, and orange, respectively. The closest distances between flavin rings and nonhydrogen atoms of side chains are shown in angstroms. (C) Steady-state absorption spectra of flavin species in different systems. FAD•− in GOX was generated by photoreduction in the presence of ethylenediaminetetraacetic acid at pH 10.1, and the spectrum of FADox in GOX was measured at pH 10.1. Spectra of FAD•− in DAAO and RF•− in solution were reproduced from published data (26, 42). The other spectra were measured, as described in the SI Appendix, Methods. For all the spectra, the reported values of extinction coefficients were used (26, 27, 30, 31). Dashed lines indicate the maxima of excitation pulses in the time-resolved spectroscopic measurements. (D) HOMO and LUMO energy levels of FAD•− calculated in vacuum and in GOX at the ωB97X-D3/ma-def2-TZVP level.