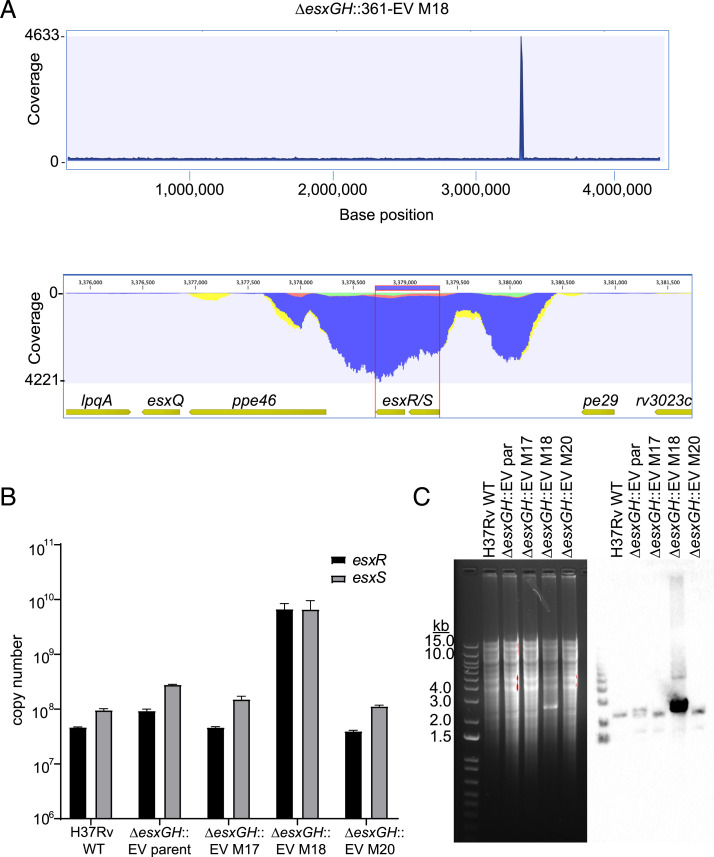
Fig. 5.
Massive in vivo amplification of a portion of the esxRS locus in a ΔesxGH strain. (A) Whole-genome Illumina sequencing read coverage (read length 150 bp) for strain H37Rv ΔesxGH::361-EV M18. (Upper) Read coverage across the bacterial genome. (Lower) Zoom-in on an ∼3-kb region that includes the esxS-esxR genes and flanking sequences, demonstrating that this region corresponds to the sharp spike in coverage seen in the Upper panel. (B) qPCR was performed in order to determine the esxR and esxS copy numbers in equivalent amounts of genomic DNA from H37Rv WT, ΔesxGH::361-EV (the parental strain used to inoculate mice), and the three isolates recovered from mice #17, #18, and #20. The data represent the mean esxR and esxS copy numbers, determined in triplicate and calculated from standard curves. Error bars represent the SD. (C, Left) An ethidium bromide-stained agarose gel separating BamHI-digested genomic DNA from the indicated stains. (Right) The results of a Southern blot of the samples probed for esxS-esxR.