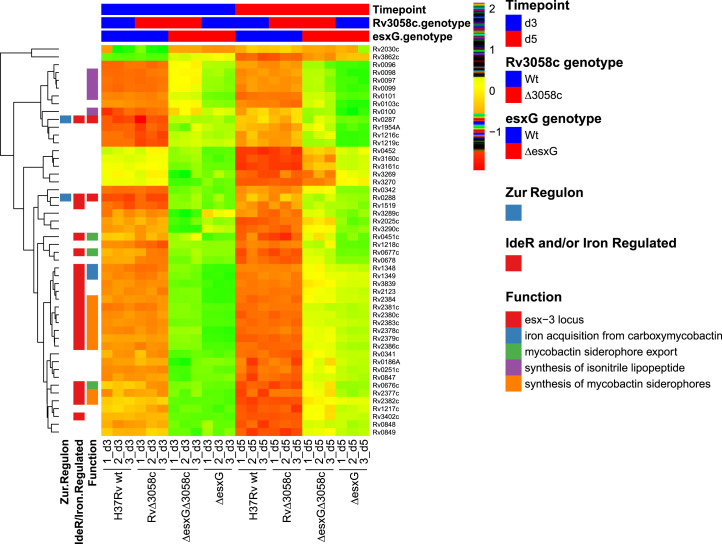
Fig. 7.
Transcriptomic analysis using RNA-seq reveals a metal starvation signature for the ΔesxG mutant grown in 7H9 medium, which is partially reversed by deleting rv3058c. The hierarchical clustering heatmap represents color-coded expression levels of the top 50 significant DEGs identified when comparing ΔesxG with H37Rv WT at day 5 of growth in 7H9 medium, based on the log2 fold-change values (false-discovery rate < 5%) as determined by RNA-seq analysis. Each row of the heat map represents a different gene as indicated to the right of the map and each column represents an individual sample as indicated below the map. The color and intensity of each cell are related to the normalized gene expression values, with green or red colors indicating up or down-regulation respectively and reordered according to the hierarchical clustering results. Several gene sets in functional categories relevant to this study are denoted by the colors in the legend on the right (Function). Genes that have previously been defined as being regulated by IdeR/iron or as belonging to the Zur regulon are indicated in the left-most columns. The groupings regarding time point, as well as rv3058c and esxG genotypes (WT or mutant) are indicated above the heat map.