Abstract
Spinal cord impairment involving motor neuron degeneration and demyelination can cause lifelong disabilities, but effective clinical interventions for restoring neurological functions have yet to be developed. In early spinal cord development, neural progenitors of the motor neuron (pMN) domain, defined by the expression of oligodendrocyte transcription factor 2 (OLIG2), in the ventral spinal cord first generate motor neurons and then switch the fate to produce myelin-forming oligodendrocytes. Given their differentiation potential, pMN progenitors could be a valuable cell source for cell therapy in relevant neurological conditions such as spinal cord injury. However, fast generation and expansion of pMN progenitors in vitro while conserving their differentiation potential has so far been technically challenging. In this study, based on chemical screening, we have developed a new recipe for efficient induction of pMN progenitors from human embryonic stem cells. More importantly, these OLIG2+ pMN progenitors can be stably maintained for multiple passages without losing their ability to produce spinal motor neurons and oligodendrocytes rapidly. Our results suggest that these self-renewing pMN progenitors could potentially be useful as a renewable source of cell transplants for spinal cord injury and demyelinating disorders.
Keywords: human embryonic stem cells, neural differentiation, chemical approach, self-renewing, progenitors of motor neurons, oligodendrocytes
Introduction
During early spinal cord development in vertebrate embryos, neural progenitors of the motor neuron (pMN) domain in the ventral spinal cord first generate different subtypes of motor neurons (directly responsible for skeletal muscle movements) and then switch abruptly to the production of oligodendrocyte (OL) precursor cells (OPCs), which subsequently give rise to myelin-forming OLs in the spinal cord. pMN progenitors are marked by the expression of OL transcription factor 2 (OLIG2) and are known to contribute to all the motor neurons and most OPCs of the spinal cord in mice (Park et al., 2004; Jakovcevski and Zecevic, 2005; Ligon et al., 2006; Lu et al., 2015). In humans, spinal cord conditions featuring motor neuron and myelin degeneration, such as spinal cord injury, spinal muscular atrophy, amyotrophic lateral sclerosis, and multiple sclerosis (Franklin and Ffrench-Constant, 2008; Burghes and Beattie, 2009; Ling et al., 2013; Tosolini and Sleigh, 2017; Borghi et al., 2019; Saladini et al., 2020), can cause life-changing disabilities with devastating consequences, but effective treatments for restoring spinal cord neurological functions have yet to be found (Burghes and Beattie, 2009; Borghi et al., 2019; Saladini et al., 2020). With the advance of stem cell research, stem cell-based cell therapy has become an emerging strategy for combating these incurable diseases.
Generation of motor neurons or OLs from mouse and human pluripotent stem cells (PSCs), including induced PSCs (iPSCs) and embryonic stem cells (ESCs), has been intensively investigated in the past decades (Brustle et al., 1999; Billon et al., 2002; Wichterle et al., 2002; Keirstead et al., 2005; Li et al., 2005; Hu et al., 2009; Liu et al., 2011; Douvaras and Fossati, 2015; Du et al., 2015). The key rationale for these studies is mimicking developmental principles in vitro. For example, spinal motor neurons can be generated from ESCs by stepwise induction, including (i) neural induction through inhibition of transforming growth factor β (TGF-β) and bone morphogen protein (BMP) signaling pathways, (ii) caudalization and ventralization through exposure to retinoic acid (RA) and sonic hedgehog (SHH), and (iii) final neuronal maturation (Hu and Zhang, 2009; Davis-Dusenbery et al., 2014; Du et al., 2015). The motor neurons generated from PSCs have been proven to resemble bona fide motor neurons in many aspects, including electrophysiological properties and the ability to form neuromuscular junctions with muscles (Davis-Dusenbery et al., 2014). The generation of the OL lineage follows a similar approach, including neural induction and ventralization, OPC induction and expansion, and final OL maturation (Czepiel et al., 2011; Wang et al., 2013; Douvaras et al., 2014; Goldman and Kuypers, 2015). PSC-derived OPCs have been found able to effectively remyelinate axons following transplantation and have significant therapeutic effects in rodent and primate models of demyelination (Wang et al., 2013; Douvaras et al., 2014; Piao et al., 2015; Thiruvalluvan et al., 2016).
Despite numerous successes, at present, generating motor neurons or OLs from PSCs is usually an extremely time-consuming process, and obtaining self-renewing intermediate multipotent progenitors with differentiation potential for both motor neuron and OL production remains a challenging task. Given their distinct multipotencies, pMN progenitors have the potential to be a new renewable cell source for cell replacement therapy in relevant medical conditions, especially spinal cord injury. In the current study, we unveil a new chemically defined culture system, which is developed based on the known signals for pMN domain formation and fate choice in spinal cord development. Using this culture system, human ESCs (hESCs) can be induced to rapidly and efficiently differentiate into self-renewing OLIG2+ pMN progenitors that can be long-term maintained and are able to produce both motor neurons and OPCs/OLs in a short time frame. These hESC-derived pMN progenitors provide a new tool for cell therapy, disease modeling, and drug screening.
Results
Generation of pMN progenitors from hESCs
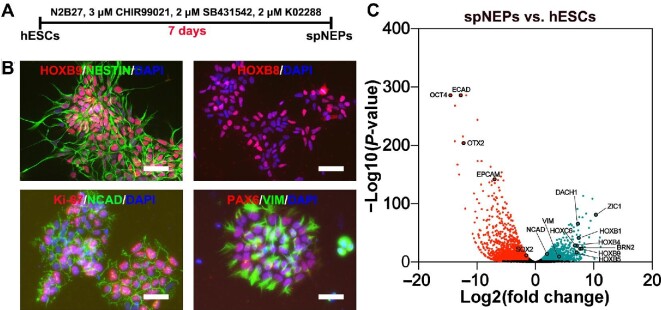
To generate pMN progenitors, we first induced hESCs (Hues9 and H1 cell lines), cultured under feeder-free conditions on Matrigel in E8 medium, to differentiate into spinal cord neuroepithelial progenitors (spNEPs). When the cells reached 30% confluence, small molecules, including 3 μM glycogen synthase kinase 3 (GSK3) inhibitor CHIR99021, 2 μM TGF-β receptor inhibitor SB431542, and 2 μM BMP receptor inhibitor K02288, were added to the medium to induce posterior neural differentiation (Figure 1A). After 1-week induction, differentiation into neuroepithelia was achieved as evidenced by the expression of NESTIN, N-cadherin (NCAD), PAX6, and vimentin (VIM) (Figure 1B). Moreover, homeobox proteins HOXB8 and HOXB9 were also expressed in these neuroepithelial progenitors (NEPs), pointing to spinal cord identity (Figure 1B). These caudalized cells were highly proliferative and stained positive for Ki-67 (Figure 1B). RNA sequencing (RNA-seq) analysis further confirmed both NEP (NESTIN+/NCAD+/VIM+/BRN2+/ZIC1+) and spinal cord (HOXB1+/HOXB4+/HOXB5+/HOXC6+/HOXB9+) identities for these spNEPs (Figure 1C).
Figure 1.
The induction of spNEPs from hESCs. (A) Schematic representation of spNEP induction. (B) Representative immunocytochemistry images of spNEPs. The expression of HOXB9/HOX8/Ki-67/PAX6 (red) and NESTIN/NCAD/VIM (green) was detected in spNEPs. Nuclei were counterstained with DAPI. (C) Volcano plot of differential gene expression by RNA-seq in hESCs and spNEPs. PSC-enriched genes are in red; spNEP-enriched genes are in light blue. Scale bar, 50 μm.
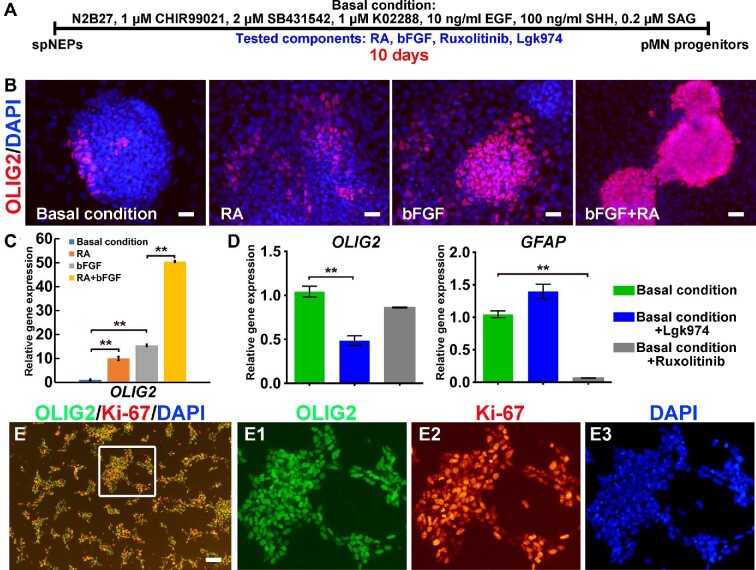
Previously, we demonstrated successful induction of self-renewing primitive neural stem cells from hESCs using CHIR99021, SB43142, and human leukemia inhibitory factor (LIF) (Li et al., 2011). Based on previous studies, as a starting point, we devised basal conditions where culture medium was supplemented with a combination of well-established neural stem cell mitogens (CHIR99021, SB4315422, K02288, and epidermal growth factor [EGF]) and ventralizing morphogens (SHH and smoothened agonist [SAG]), aiming at generating self-renewing OLIG2+ pMN progenitors (Figure 2A). Since only a small number of OLIG2+ cells could be induced from spNEPs after 10 days by culturing with this basal medium alone (Figure 2B), we further searched for additional ingredients in order to promote the generation of pMN progenitors. We tested several small molecules that are known to modulate essential signaling pathways, such as RA, basic fibroblast growth factor (bFGF), and Wnt and Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) signaling, in neural patterning and cell fate determination by performing iterative chemical screening as reported previously (Lv et al., 2015; Sun et al., 2019). By evaluating OLIG2 expression with quantitative reverse transcription‒polymerase chain reaction (qRT‒PCR) and immunocytochemistry, we first identified RA and bFGF being able to increase OLIG2 expression and the number of OLIG2+ cells, especially when combined together (Figure 2B and C). Fluorescence-activated cell sorting (FACS) analysis demonstrated that the proportion of OLIG2+ cells rose from 3.7% under basal conditions to 9.17%, 44%, and 80.4% with the addition of RA, bFGF, and RA combined with bFGF, respectively (Supplementary Figure S1). Notably, treating the cells with ruxolitinib, an inhibitor of JAK2/STAT3 signaling, had no significant impact on OLIG2 expression but dramatically decreased the expression of the astrocyte marker glial fibrillary acidic protein (GFAP) (Figure 2D), which is consistent with the favorable role of JAK2/STAT3 signaling in astrocyte fate determination (Yanagisawa et al., 1999). In addition, Lgk974, a Wnt inhibitor, led to reduced OLIG2 expression (Figure 2D). Building on these results, we established a new formula consisting of CHIR99021, SB431542, K02288, EGF, SHH, SAG, RA, bFGF, and ruxolitinib for the induction of OLIG2+ pMN progenitors. This new cocktail effectively promoted the conversion from spNEPs into proliferating OLIG2+ pMN progenitors, which also expressed Ki-67 (Figure 2E).
Figure 2.
The induction of OLIG2+ pMN progenitors from spNEPs. (A) Schematic representation of OLIG2+ pMN progenitor induction. Additional components were tested in combination with the basal medium by reiterative chemical screening for their effect on the induction. (B–D) The impact of RA, bFGF, ruxolitinib, and Lgk974 on OLIG2 expression as analyzed by immunocytochemistry (B) and qRT‒PCR (C and D). spNEPs were cultured in the basal medium or in combination with RA and/or bFGF for generation of OLIG2+ (red) progenitors (B). Gene expression (C and D) was relative to GAPDH (data from three experiments, mean ± SEM). (E) Efficient induction of OLIG2+ pMN progenitors from spNEPs by the basal medium supplemented with RA, bFGF, and ruxolitinib. E1, E2, and E3 show the boxed area in E at a higher magnification. Scale bar, 25 μm (B) and 100 μm (E). **P < 0.01.
Self-renewal of pMN progenitors
Despite the efficient induction of pNM progenitors that expressed OLIG2, NESTIN, and HOXB9 (Figure 3A and B), routine subculture of these cells resulted in a gradual loss of OLIG2-expressing cells but an increase in the number of S100β+ cells (Supplementary Figure S2), suggesting a shift toward astrocytic lineage.
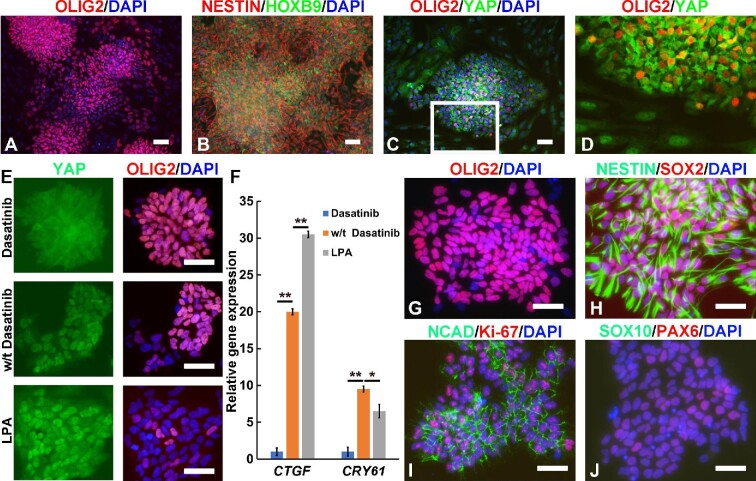
Figure 3.
Dasatinib maintains stable expression of OLIG2 by inhibiting YAP signaling in OLIG2+ pMN progenitors. (A‒D) Immunocytochemistry images of pMN progenitors cultured in the basal medium supplemented with RA, bFGF, and ruxolitinib. A higher-magnification image of the boxed area in C is shown in D. (E) Immunocytochemistry images of OLIG2+ pMN progenitor-derived single-cell clones treated with/without dasatinib or LPA. (F) qRT‒PCR analysis of the expression of YAP target genes CTGF and CRY61 in OLIG2+ pMN progenitors treated with/without dasatinib or LPA. (G‒J) Immunocytochemistry images of pMN progenitors (at passage 10) cultured in the basal medium supplemented with RA, bFGF, ruxolitinib, and dasatinib. Scale bar, 100 μm (A‒D) and 80 μm (E and G‒J). **P < 0.01, *P < 0.05.
Interestingly, we frequently noticed that OLIG2– cells usually contoured the OLIG2+ cell clusters, which was reminiscent of our previous observation that hepatocytes are more profoundly de-differentiated at the border of an epithelial cluster due to mechanical tension-induced YAP activation (Sun et al., 2019). Indeed, immunocytochemistry results showed that OLIG2+ cells with cytoplasmic retention of YAP were surrounded by OLIG2– cells, in which YAP protein accumulated in the nucleus (Figure 3C and D). These data implied that the loss of OLIG2 expression might be triggered by YAP activation. To evaluate this hypothesis, we generated single-cell colonies and treated the cells with dasatinib, an Src family tyrosine kinase inhibitor that can inhibit YAP activation. As expected, addition of dasatinib prevented YAP nuclear localization while effectively sustaining OLIG2 expression (Figure 3E). In contrast, lysophosphatidic acid (LPA) treatment induced YAP activation as reported and substantially disrupted OLIG2 expression (Figure 3E). qRT‒PCR analysis confirmed that dasatinib repressed the expression of YAP target genes such as CTGF and CRY61, whereas LPA activated CTGF expression (Figure 3F). Therefore, dasatinib was added to our recipe for maintaining long-term self-renewal of OLIG2+ pMN progenitors. Notably, in the presence of dasatinib, OLIG2+ pMN progenitors could be stably maintained for >20 passages without significant loss of OLIG2 expression (Figure 3G). pMN progenitors were routinely passaged 1:3 every 2 days and could be cultured for >20 passages with no obvious loss of proliferative capacity as demonstrated by FACS analysis of OLIG2+ pMN progenitors (92% and 86.3% of OLIG2+ cells at passage 3 and passage 21, respectively, Supplementary Figure S1). Cell cycle analysis also revealed that pMN progenitors at passage 3 and passage 21 had similar distributions across three major phases of the cycle (Supplementary Figure S3). pMN progenitors expressed neural stem cell markers, including NESTIN, SOX2, NCAD, and PAX6 (Figure 3H‒J), and were homogenously positive for the cell proliferation marker Ki-67 (Figure 3I). RNA-seq analysis demonstrated upregulation and stable expression of a panel of genes characteristic of pMN progenitors, including OLIG2, OLIG1, NKX6.1, and NKX6.2, in these pMN progenitors, whereas the expression of IRX3, a marker of domains dorsal to pMN, was significantly downregulated (Supplementary Figure S4).
To confirm the effect of Src inhibition on the maintenance of pMN progenitors, we tested saracatinib, another Src family tyrosine kinase inhibitor with higher specificity, and found that it had the same effect as dasatinib. Similar to using dasatinib, long-term cultured pMN progenitors using saracatinib exhibited characteristic phenotypes of pMN progenitors, including expression of OLIG2, HOXB9, and Ki-67 (Supplementary Figure S5).
pMN progenitors can differentiate into spinal motor neurons
During spinal cord development, pMN progenitors are multipotent, able to give rise to both spinal motor neurons and OPCs. Therefore, we investigated whether these hESC-derived self-renewing pMN progenitors retained this multipotency. However, motor neuron differentiation medium comprising brain-derived neurotrophic factor (BDNF), glial cell-derived neurotrophic factor (GDNF), RA, and SAG did not induce homogenous neuronal differentiation; instead, the culture deteriorated with overgrowth of astrocyte-like cells (Supplementary Figure S6A). qRT‒PCR confirmed the upregulation of S100β (astrocyte marker) expression (Supplementary Figure S6B).
Previous studies demonstrate that GSK3 regulates the equilibrium between neurogenesis and gliogenesis and GSK3 inhibition favors neurogenesis (Singh et al., 2018). To promote motor neuron differentiation, we titrated CHIR99021 (ranging from 0.5 to 3 μM) to assess the impact on neurogenesis from pMN progenitors. After 1-week culture, we found that the expression of neuronal marker neurogenin-2 (NGN2), which is responsible for neurogenesis, was dramatically enhanced with the increase of CHIR99021 concentration, whereas the expression of nuclear factor IA (NFIA), a master regulator of astrocyte development, was markedly downregulated toward the high end of CHIR99021 concentration (Supplementary Figure S7). OLIG2 showed no obvious change when the CHIR99021 concentration was in the 1‒3 μM range, but lowering the CHIR99021 concentration to 0.5 μM could compromise OLIG2 expression. The expression of the motor neuron marker vesicular acetylcholine transporter (vAChT) showed a clear upward trend with increasing CHIR99021 concentration (Supplementary Figure S7).
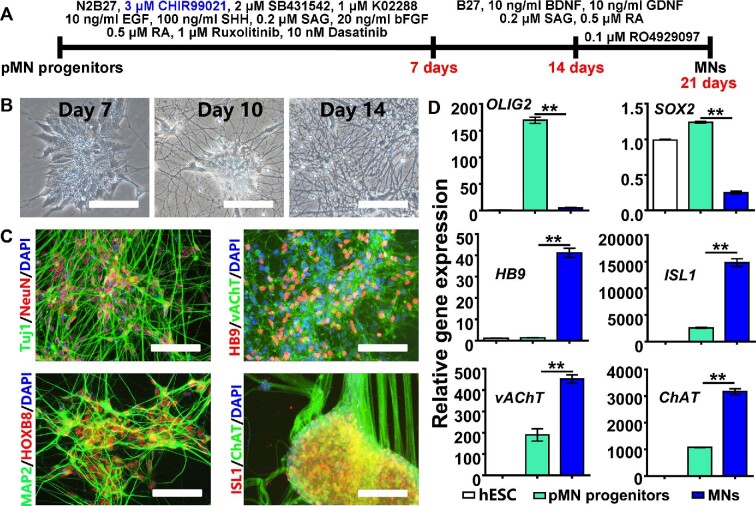
Based on the above results, we optimized our motor neuron induction protocol and designed two-step induction: (i) pMN progenitors were first cultured for a week in self-renewing medium with 3 μM CHIR99021; (ii) the culture continued in motor neuron differentiation medium for 2 weeks with the γ-secratase inhibitor RO4929097 being added in the last week to accelerate cell maturation (Figure 4A). Under these optimized conditions, pMN progenitors could rapidly and homogenously differentiate into a neuronal population (Figure 4B). These induced neurons expressed neuronal markers such as Tuj1, microtubule associated protein 2 (MAP2), mature neuron-specific nuclear protein NeuN (neuronal nuclei), as well as the spinal cord neuron marker HOXB8 (Figure 4C). Moreover, these neurons exhibited motor neuron-specific gene expression, including the expression of homeobox protein HB9, vAChT, ISL LIM homeobox 1 (ISL1), and choline acetyltransferase (ChAT) (Figure 4C), which was further confirmed by qRT‒PCR (Figure 4D). Importantly, early- and late-passage pMN progenitors demonstrated similar potentials for spinal motor neuron induction (Supplementary Figure S8).
Figure 4.
Induction of spinal motor neurons from pMN progenitors at passage 15. (A) Schematic representation of motor neuron induction from pMN progenitors. (B) Phase-contrast images of cells at different time points of differentiation induction. (C and D) Neuron- and motor-neuron-specific gene expression after differentiation induction was analyzed by immunocytochemistry (C) and qRT‒PCR (D). MN, motor neuron. Scale bar, 100 μm. **P < 0.01.
pMN progenitors can differentiate into OLs
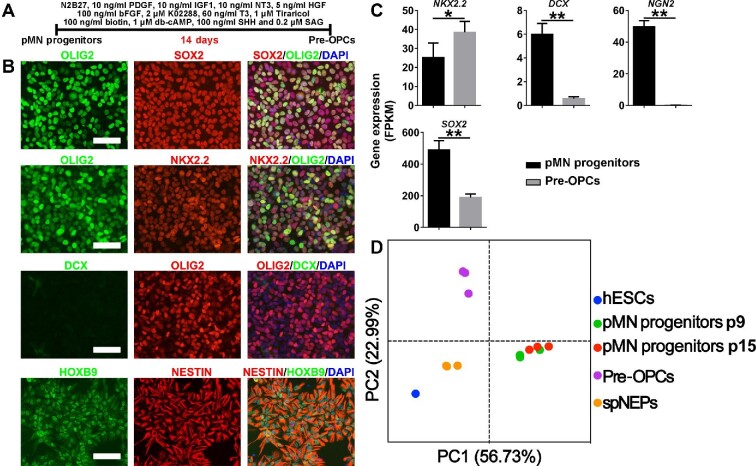
Next, we examined the differentiation potential of pMN progenitors toward OLs. After 2-week culture in OPC induction medium (Figure 5A), although the cells still expressed neural progenitor markers such as OLIG2, SOX2, and NESTIN (Figure 5B), the expression of proneuronal factors DCX and NGN2 was greatly downregulated (Figure 5B and C). The expression of NK2 homeobox 2 (NKX2.2) was upregulated (Figure 5B) and NKX2.2 protein was coexpressed with OLIG2 (Figure 5C), which is a hallmark of OPC program initiation (Zhou et al., 2001), suggesting a stage of pre-OPCs. Principal-component analysis (PCA) of RNA-seq data showed that samples belonging to the same differentiation stage clustered together, which was also supported by hierarchical clustering of fragments per kilobase of transcript per million mapped reads (FPKM) values (Supplementary Figure S9A and B). For example, pMN progenitors from passage 9 and passage 15 were remarkably similar (Figure 5D; Supplementary Figure S9A and B). Gene set enrichment analysis (GSEA) on RNA-seq data from pMN progenitors versus spNEPs showed gene enrichment for amyotrophic lateral sclerosis and axon guidance in pMN progenitors (Supplementary Figure S9C). KEGG pathway enrichment analysis revealed that genes representing axon guidance, cholinergic synapse, and GABAergic synapse were significantly enriched in pMN progenitors compared with spNEPs (Supplementary Figure S9D).
Figure 5.
Induction of pre-OPCs from passage 15 pMN progenitors. (A) Schematic representation of pre-OPC induction from pMN progenitors. (B and C) The expression of neural progenitor markers (OLG2, SOX2, and NESTIN) as well as molecular indicators of neurogenesis (NGN2, DCX, and HOXB9) and oligodendrogenesis (OLIG2/NKX2.2) was analyzed by immunocytochemistry (B) and RNA-seq (C). (D) PCA for gene expression of hESCs, spNEPs, pMN progenitors, and pre-OPCs. Each dot is one biological replicate. Scale bar, 100 μm. **P < 0.01, *P < 0.05.
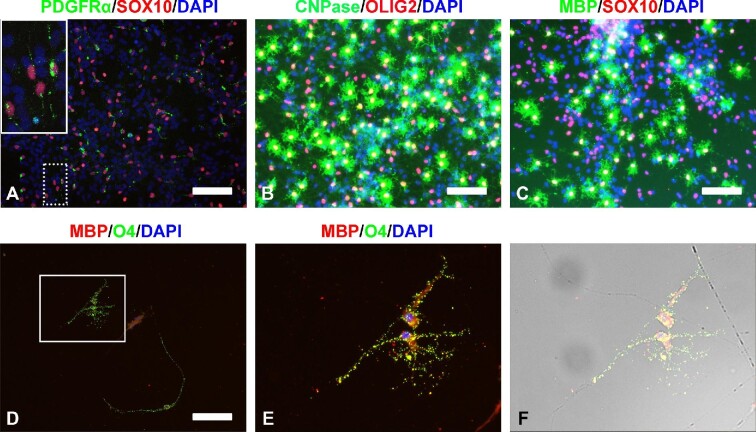
To promote OPC maturation, growth factors and small molecules, including platelet derived growth factor (PDGF), insulin-like growth factor 1 (IGF1), neurotrophin 3 (NT3), hepatocyte growth factor (HGF), SHH, SAG, and K02288, were removed from OPC induction medium, and after another 2-week culture, bipolar SOX10+/PDGFRα+ OPCs were obtained (Figure 6A). We further proved that with two more weeks of culture, these OPCs could generate multipolar ‘spidery’ OLs positive for mature OL markers myelin basic protein (MBP) and 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase) as well as OL lineage markers OLIG2 and SOX10 (Figure 6B and C). Thus, after a total of 6 weeks of culture, the differentiation from pMN progenitors into mature OLs was completed. Furthermore, 2 weeks after seeding the induced OPCs on poly(lactic-co-glycolic acid) (PLGA) nanofibers, MBP+/O4+ mature OLs could be seen enwrapping the nanofibers with myelin (Figure 6D‒F), indicating these OPCs’ ability to produce myelin-forming OLs. Interestingly, late-passage pMN progenitors seemed more susceptible to OL induction (Supplementary Figure S10).
Figure 6.
Induction of OLs from pre-OPCs derived from passage 15 pMN progenitors. (A) Immunocytochemistry images of SOX10+/PDGFRα+ bipolar OPCs obtained from pre-OPCs after a 2-week induction. The inset in A is a higher-magnification view of the area in the dotted white box. (B and C) Immunocytochemistry images of mature OLs expressing CNPase/OLIG2 (B) and MBP/SOX10 (C) after 2 weeks of induction from bipolar OPCs. (D‒F) Images of MBP+/O4+ mature OLs enwrapping PLGA nanofibers with myelin. OPCs were seeded on PLGA nanofibers and cultured for 2 weeks. (E and F) Higher-magnification images of the boxed area in D. F is derived from E superimposed on the corresponding phase-contrast image. Scale bar, 100 μm (A and D) and 80 μm (B and C).
Taken together, the above data suggest that these long-term cultured self-renewing pMN progenitors can retain the potential to differentiate into both spinal motor neurons and OPCs/OLs within a short time span, which makes them a promising source of cell transplants, especially for treating spinal cord injury.
Discussion
In recent years, stem cell-based cell therapy has provided a new approach to treating neurological conditions and has been widely investigated. Human PSCs (hPSCs), i.e. human iPSCs and hESCs, are theoretically a renewable source of any type of cells, and tremendous progress has been made in the recent decades in inducing hPSC differentiation toward motor neurons and OLs. However, so far, the production of motor neurons or OPCs involves very lengthy procedures. Given their ability to generate motor neurons and OPCs, pMN progenitors have the potential to be used directly as cell transplants for certain spinal cord conditions, especially spinal cord injury, or as a source of OPC transplants to treat demyelinating disorders. To date, there has been limited success in producing pMN progenitors from hESCs and a major obstacle encountered is stable expansion of pMN progenitors in vitro. In this study, equipped with our previous success in expanding hESC-derived primitive neural stem cells (Li et al., 2011), we used a reiterative screening strategy to successfully establish chemically defined conditions for maintaining the self-renewal of hESC-derived OLIG2+ pMN progenitors over a high number of passages (>20) (Figure 3). These self-renewing pMN progenitors gave rise to HB9+/ChAT+ motor neurons and MBP+ mature OLs in 3 and 6 weeks, respectively (Figures 4‒6), much faster than inducing hPSCs to differentiate into these specialized cells. For a renewable cell source and immediate progenitors of motor neurons and OPCs, it will be intriguing to find out how pMN progenitors fulfill their potential in vivo in suitable animal models in the future.
Although we can successfully maintain pMN progenitors in a stable, self-renewing state in vitro, a couple of questions regarding the molecular mechanisms involved still need to be further investigated. For instance, in this study, we found that the Src inhibitor dasatinib was crucial for stable OLIG2 expression, possibly through YAP inhibition (Figure 3), but the mechanisms underlying the regulation of OLIG2 expression by YAP signaling remain obscure. In addition, to tackle astrocyte overgrowth, a common obstacle to neuronal differentiation from neural progenitors in vitro, we temporarily increased the CHIR99021 concentration from 1 to 3 μM, which dramatically upregulated the expression of proneuronal factor NGN2 and enhanced neurogenesis from pMN progenitors (Supplementary Figures S4 and S5). Previous studies showed that GSK3 could phosphorylate and regulate NGN2 functions with little impact on NGN2 expression in neocortical neurogenesis and spinal cord motor neuron development (Ma et al., 2008; Li et al., 2012). How exactly GSK3 regulates NGN2 expression and function and controls pMN progenitor fate determination still needs further elucidation. It is also worth noting that although short-term treatment (e.g. a week) of CHIR99021 at 3 μM did not influence OLIG2 expression of pMN progenitors (Supplementary Figure S5), long-term passaging of pMN progenitors using 3 μM CHIR99021 could diminish OLIG2 expression (data not shown).
Chemical approaches using small molecules have been proven to be especially useful in modulating cell fate in stem cell research (Li et al., 2013). Following clues found in spinal cord development, our generation of self-renewing multipotent pMN progenitors by stepwise induction using a combination of growth factors and compounds once again demonstrates the power of chemical approaches in manipulating stem cell fate. Similar strategies can be explored in generating other clinically relevant self-renewing somatic stem cells or progenitors from hPSCs, as well as in improving the differentiation efficiency of certain specialized cells both in vitro and in vivo.
Materials and methods
hESC culture and generation of self-renewing pMN progenitors
hESCs of Hues9 cell line (passages 20‒30) and H1 cell line (passages 50‒60) were cultured in E8 medium on plates coated with 2% Matrigel as described previously (Yao et al., 2006). The cells were regularly passed using TrypLE. hESCs at ∼30% confluence were cultured in differentiation medium containing Dulbecco’s modified Eagle’s medium/nutrient Ham’s mixture F-12 (DMEM/F-12) (1× N2, 1×B27) supplemented with 3 μM CHIR99021, 2 μM SB431542, and 2 μM K02288 for 1 week to obtain spNEPs.
spNEPs were then cultured in ventralization and expansion medium containing DMEM/F12 (1 × N2, 1×B27) supplemented with 1 μM CHIR99021, 2 μM SB431542, 1 μM K02288, 0.2 μM SAG, 100 ng/ml SHH, 10 ng/ml EGF, 20 ng/ml bFGF, 0.5 μM RA, 10 nM dasatinib, and 100 nM ruxolitinib for ∼10 days and robust generation of OLIG2+ pMN progenitors could be obtained. pMN progenitors were routinely passaged 1:3 every 2 days and could be cultured for >20 passages on Matrigel-coated 6-well plates without obviously losing proliferative capacity. Overnight treatment with the myosin II ATPase inhibitor (1 μM blebbistatin) was used to improve cell survival after a cell passage during neural induction but not required for routine culture of OLIG2+ pMN progenitors.
All tissue culture products were obtained from Invitrogen except where mentioned.
Motor neuron differentiation from self-renewing pMN progenitors
OLIG2+ pMN progenitors were cultured on Matrigel-coated 6-well plates. When cells reached ∼30% confluence, the CHIR99021 concentration in the medium was raised to 3 μM for 1-week treatment, during which cells were routinely split 1:3 by using TrypLE, and then the treated cells were reseeded on Matrigel-coated plates. When the cell confluence reached 30%, terminal differentiation was induced by switching to motor neuron differentiation medium containing neural basal medium (1× B27 plus) supplemented with 10 ng/ml BDNF, 10 ng/mGDNF, 0.2 μM SAG, and 0.5 μM RA. After 1 week of treatment, when most cells differentiated into neurons, 0.1 μM γ-secratase inhibitor RO4929097 was added into the medium for another week of culture to accelerate cell maturation.
OL induction from self-renewing pMN progenitors
OLIG2+ neural progenitors were cultured on Matrigel-coated 6-well plates. After reaching 30% confluence, these cells were placed in glial progenitor induction medium containing DMEM/F12 (1× N2, 1× B27) supplemented with 10 ng/ml PDGF, 10 ng/ml IGF1, 10 ng/ml NT3, 5 ng/ml HGF, 100 ng/ml bFGF, 2 μM K02288, 60 ng/ml triiodo-L-thyronine (T3), 1 μM tiraricol (metabolite of T3), 100 ng/ml biotin, 1 μM dibutyryl cyclophosphamide (db-cAMP), 100 ng/ml SHH, and 0.2 μM SAG for 1 week of culture to get pre-OPCs. Cells were routinely split 1:3 while being induced. After removal of reagents PDGF, IGF1, NT3, HGF, and K02288 from the medium, the cells were further cultured for 1 week to obtain bipolar OPCs, which were then dissociated, reseeded on a laminin-coated surface, and cultured with terminal differentiation medium containing DMEM/F12 (1× N2, 1× B27) supplemented with 60 ng/ml T3, 1 μM tiraricol, 100 ng/ml biotin, and 1 μM db-cAMP for ∼2‒4 weeks for cell maturation.
qRT‒PCR and RNA-seq
Total RNA was extracted using TRIzol (Invitrogen). Reverse transcription was performed with 1 μg RNA using PrimeScript™ RT Master Mix (TaKaRa). The primers used are shown in Supplementary Table S1. qRT‒PCR was carried out using iQ SYBR Green Supermix (Bio-Rad). Gene expression was quantified by normalizing to glyceraldehyde-phosphate dehydrogenase (GAPDH) expression. For RNA-seq analysis, total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion). The samples with an RNA integrity number (RIN) >7, measured using the Agilent 2100 Bioanalyzer (Agilent Technologies), were subjected to sequencing analysis. The cDNA libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) and sequenced with the Illumina HiSeqTM 2500 system. Raw data were processed using Trimmomatic. Reads containing poly-N and low-quality reads were removed to obtain clean reads, which were then mapped to the reference genome using HISAT2. The FPKM value of each gene was calculated using Cufflinks, and the read counts of each gene were obtained with the htseq-count script. Differentially expressed genes (DEGs) were identified using DESeq. P-value <0.05 and fold change >2 or <0.5 were set as the threshold for significantly differential expression.
Immunocytochemistry
For immunocytochemistry analysis, cells were fixed in 4% paraformaldehyde for 5 min at room temperature, washed three times with PBST (phosphate-buffered saline containing 0.2% Triton X-100), and incubated in blocking buffer consisting of PBST with 5% normal donkey serum (Jackson ImmunoResearch Laboratories) for 30 min at room temperature. The cells were then incubated with primary antibodies (listed in Supplementary Table S2) in blocking buffer overnight at 4°C. Next, the cells were washed three times with PBST and incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen, 1000×) in PBST for 1 h at room temperature. Nuclei were visualized by 4′,6-diamidino-2-phenyliindole (DAPI; Sigma-Aldrich) staining. Images were captured using a Nikon Eclipse 50i microscope.
FACS
For indirect flow cytometry, cells were dissociated with TrypLE into single cells. BD Cytofix/Cytoperm Kit (BD Biosciences) was used for fixation, permeabilization, and washing. Then, the fixed cells were incubated with goat anti-OLIG2 antibody (R&D, 1:1000) on ice for 1 h and washed three times. Cells were then incubated with Alexa Fluor 488 F(ab’)2 donkey anti-goat (Invitrogen, 1:1000) for 30 min on ice and washed three times. Cells directly incubated with secondary antibody were used as control. Cells were analyzed using BD FACSVerse (BD Biosciences).
Statistical analysis
Statistical analysis was performed using Microsoft Excel and GraphPad Prism Software. Statistical significance (P-value) between two groups and among multiple groups was analyzed by unpaired t-test and one-way analysis of variance followed by Tukey's multiple comparisons test, respectively. P < 0.05 was considered to be statistically significant.
Supplementary Material
Contributor Information
Guan-Yu Zhang, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Zhu-Man Lv, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Hao-Xin Ma, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Yu Chen, Changhai Hospital, Second Military Medical University, Shanghai 200433, China.
Yuan Yuan, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Ping-Xin Sun, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Yu-Qi Feng, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Ya-Wen Li, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Reproductive Medicine, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Wen-Jie Lu, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Reproductive Medicine, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Yu-Dong Yang, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Reproductive Medicine, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Cheng Yang, Wolfson Institute for Biomedical Research, University College London, Gower Street, London WC1E 6BT, UK.
Xin-Lu Yu, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Chao Wang, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Shu-Long Liang, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China.
Ming-Liang Zhang, Department of Histoembryology, Genetics and Developmental Biology, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China; Shanghai Key Laboratory of Reproductive Medicine, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
Hui-Liang Li, Wolfson Institute for Biomedical Research, University College London, Gower Street, London WC1E 6BT, UK.
Wen-Lin Li, Department of Cell Biology, Second Military Medical University, Shanghai 200433, China; Shanghai Key Laboratory of Cell Engineering, Second Military Medical University, Shanghai 200433, China.
Funding
This study was supported by grants from the National Key R&D Program of China (2018YFA0107200 and 2020YFA0113101), the National Natural Science Foundation of China (81571094, 81322016, 32070866, and 31771643), the Program of Shanghai Academic Research Leader (17XD1404800), the Biotechnology and Biological Sciences Research Council (BB/S000844/1 and BB/S008934/1), Newton Advanced Fellowship (AMS-NAF1-Li), Shanghai Science and Technology Committee (19JC1413200), the program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (1710000009), and the Shanghai Key Laboratory of Reproductive Medicine.
Conflict of interest: none declared.
Author contributions: W.-L.L., H.-L.L., and M.-L.Z.: conception of study, experimental design, data analysis and interpretation, and manuscript preparation. All other authors: collection and/or assembly of data.
References
- Billon N., Jolicoeur C., Ying Q.L.et al. (2002). Normal timing of oligodendrocyte development from genetically engineered, lineage-selectable mouse ES cells. J. Cell Sci. 115, 3657–3665. [DOI] [PubMed] [Google Scholar]
- Borghi S.M., Fattori V., Hohmann M.S.N.et al. (2019). Contribution of spinal cord oligodendrocytes to neuroinflammatory diseases and pain. Curr. Med. Chem. 26, 5781–5810. [DOI] [PubMed] [Google Scholar]
- Brustle O., Jones K.N., Learish R.D.et al. (1999). Embryonic stem cell-derived glial precursors: a source of myelinating transplants. Science 285, 754–756. [DOI] [PubMed] [Google Scholar]
- Burghes A.H., Beattie C.E. (2009). Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 10, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czepiel M., Balasubramaniyan V., Schaafsma W.et al. (2011). Differentiation of induced pluripotent stem cells into functional oligodendrocytes. Glia 59, 882–892. [DOI] [PubMed] [Google Scholar]
- Davis-Dusenbery B.N., Williams L.A., Klim J.R.et al. (2014). How to make spinal motor neurons. Development 141, 491–501. [DOI] [PubMed] [Google Scholar]
- Douvaras P., Fossati V. (2015). Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 1143–1154. [DOI] [PubMed] [Google Scholar]
- Douvaras P., Wang J., Zimmer M.et al. (2014). Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Rep. 3, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.W., Chen H., Liu H.et al. (2015). Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 6, 6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R.J., Ffrench-Constant C. (2008). Remyelination in the CNS: from biology to therapy. Nat. Rev. Neurosci. 9, 839–855. [DOI] [PubMed] [Google Scholar]
- Goldman S.A., Kuypers N.J. (2015). How to make an oligodendrocyte. Development 142, 3983–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Du Z.W., Li X.J.et al. (2009). Human oligodendrocytes from embryonic stem cells: conserved SHH signaling networks and divergent FGF effects. Development 136, 1443–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Zhang S.C. (2009). Differentiation of spinal motor neurons from pluripotent human stem cells. Nat. Protoc. 4, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski I., Zecevic N. (2005). Olig transcription factors are expressed in oligodendrocyte and neuronal cells in human fetal CNS. J. Neurosci. 25, 10064–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirstead H.S., Nistor G., Bernal G.et al. (2005). Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J. Neurosci. 25, 4694–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mattar P., Zinyk D.et al. (2012). GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 32, 7791–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li K., Wei W.et al. (2013). Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sun W., Zhang Y.et al. (2011). Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl Acad. Sci. USA 108, 8299–8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.J., Du Z.W., Zarnowska E.D.et al. (2005). Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 23, 215–221. [DOI] [PubMed] [Google Scholar]
- Ligon K.L., Fancy S.P., Franklin R.J.et al. (2006). Olig gene function in CNS development and disease. Glia 54, 1–10. [DOI] [PubMed] [Google Scholar]
- Ling S.C., Polymenidou M., Cleveland D.W. (2013). Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jiang P., Deng W. (2011). OLIG gene targeting in human pluripotent stem cells for motor neuron and oligodendrocyte differentiation. Nat. Protoc. 6, 640–655. [DOI] [PubMed] [Google Scholar]
- Lu D.C., Niu T., Alaynick W.A. (2015). Molecular and cellular development of spinal cord locomotor circuitry. Front. Mol. Neurosci. 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Han Q., Chu Y.et al. (2015). Self-renewal of hepatoblasts under chemically defined conditions by iterative growth factor and chemical screening. Hepatology 61, 337–347. [DOI] [PubMed] [Google Scholar]
- Ma Y.-C., Song M.-R., Park J.P.et al. (2008). Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron 58, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.C., Shin J., Appel B. (2004). Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development 131, 5959–5969. [DOI] [PubMed] [Google Scholar]
- Piao J., Major T., Auyeung G.et al. (2015). Human embryonic stem cell-derived oligodendrocyte progenitors remyelinate the brain and rescue behavioral deficits following radiation. Cell Stem Cell 16, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladini M., Nizzardo M., Govoni A.et al. (2020). Spinal muscular atrophy with respiratory distress type 1: clinical phenotypes, molecular pathogenesis and therapeutic insights. J. Cell. Mol. Med. 24, 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Mishra A., Bharti S.et al. (2018). Glycogen synthase kinase-3β regulates equilibrium between neurogenesis and gliogenesis in rat model of Parkinson's disease: a crosstalk with Wnt and notch signaling. Mol. Neurobiol. 55, 6500–6517. [DOI] [PubMed] [Google Scholar]
- Sun P., Zhang G., Su X.et al. (2019). Maintenance of primary hepatocyte functions in vitro by inhibiting mechanical tension-induced YAP activation. Cell Rep. 29, 3212–3222.e4. [DOI] [PubMed] [Google Scholar]
- Thiruvalluvan A., Czepiel M., Kap Y.A.et al. (2016). Survival and functionality of human induced pluripotent stem cell-derived oligodendrocytes in a nonhuman primate model for multiple sclerosis. Stem Cells Transl. Med. 5, 1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosolini A.P., Sleigh J.N. (2017). Motor neuron gene therapy: lessons from spinal muscular atrophy for amyotrophic lateral sclerosis. Front. Mol. Neurosci. 10, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Bates J., Li X.et al. (2013). Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J.A.et al. (2002). Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M., Nakashima K., Taga T. (1999). STAT3-mediated astrocyte differentiation from mouse fetal neuroepithelial cells by mouse oncostatin M. Neurosci. Lett. 269, 169–172. [DOI] [PubMed] [Google Scholar]
- Yao S., Chen S., Clark J.et al. (2006). Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc. Natl Acad. Sci. USA 103, 6907–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Choi G., Anderson D.J. (2001). The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31, 791–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.