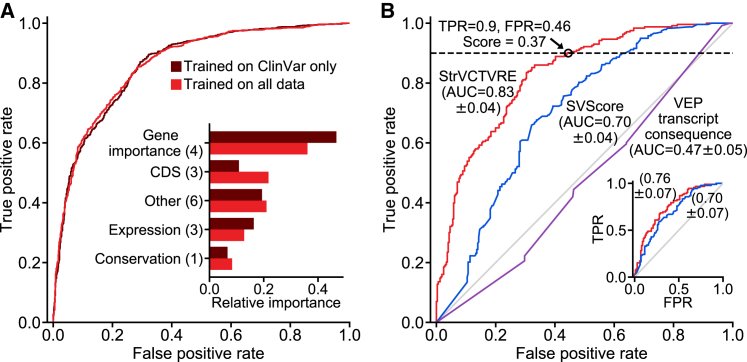
Figure 2.
Evaluation of StrVCTVRE on a held-out ClinVar test set and comparison of learned feature importances between training datasets
(A) Receiver-operating characteristic (ROC) comparing StrVCTVRE models trained on two different benign datasets: ClinVar in dark red and all data (ClinVar, SVs common to apes but not humans, and rare gnomAD SVs) in medium red. When tested only on ClinVar data, performance does not significantly differ between the two training sets. However, the feature importances (inset) of the classifier trained on all data (medium red) were more evenly distributed among feature categories. This suggests that unlabeled rare SVs and common ape SVs are a suitable benign training set.
(B) ROC comparing StrVCTVRE (red) to other methods on a held-out test set comprised of ClinVar SVs on chromosomes 1, 3, 5, and 7. Black circle indicates a StrVCTVRE score of 0.37, which we refer to as the ClinVar 90% sensitivity threshold. Inset shows performance on the same held-out test, modified so that each gene is overlapped by a maximum of one SV. AUC with 95% confidence interval is in parentheses.