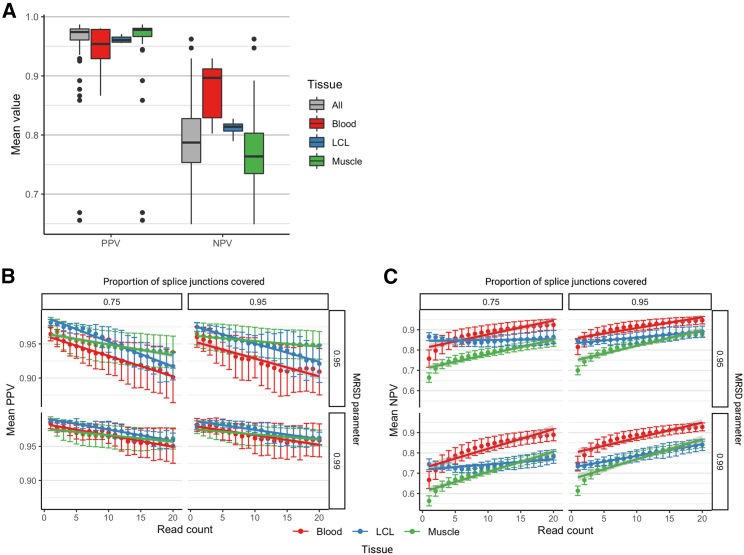
Figure 2.
Performance metrics of the MRSD model
The ability of MRSD to accurately predict levels of PanelApp disease gene coverage based on sequencing depth was tested on unseen RNA-seq datasets from blood (n = 12), LCLs (n = 4), and muscle (n = 52).
(A) The mean positive predictive values (PPVs) and negative predictive values (NPVs) averaged across all parameter combinations for each RNA-seq dataset show that the median PPV is slightly lower, and the median NPV slightly higher, for whole blood than for LCLs and skeletal muscle.
(B and C) Breakdown of (B) PPVs and (C) NPVs for the MRSD model by parameters shows that specifying an increasing desired read coverage results in a gradual decrease in PPV and increase in NPV across all tissues and parameter combinations. Dependent on parameter stringency and limiting analysis to a maximum specification of 20-read coverage, PPV predictions range from 90.1% to 98.2%, while NPV ranges from 56.4% to 94.7%. Error bars show 95% confidence interval.