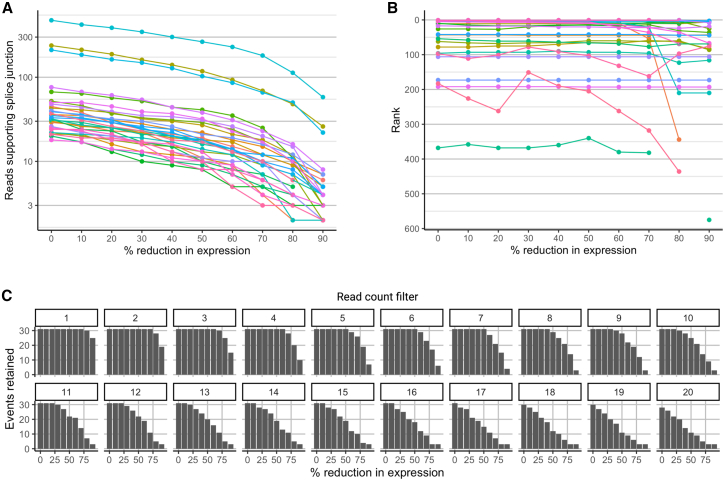
Figure 5.
Variability in expression level influences the capacity to identify mis-splicing events
Genes harboring a selection of 31 splicing events that were identified during analysis of 52 muscle-based RNA-seq datasets (and which would be identified as events of interest using a filter of normalized read count [NRC] > 0.19) were artificially downsampled to simulate variation in expression.
(A) Reduction in expression leads to an intuitive and proportional reduction in the number of reads supporting each mis-splicing event.
(B) The rank position of an event—where the event appears in a list of all splicing events in its respective sample, ordered by decreasing NRC fold change relative to controls, and placing singleton events above non-singletons—is generally consistent as expression of the gene decreases. Missing data points at the most reduced expression values are indicative of the splicing event not being identified by the applied bioinformatics pipeline.
(C) Variation in expression impacts our ability to identify events of interest when filters of read count supporting the events are enforced. When the 31 events experience a 50% reduction in expression, for instance, the application of a minimum 15-read filter leads to the exclusion of 41.9% (13/31) of events.