Figure 7.
Quantifying the power for RNA-seq to resolve variants of uncertain significance (VUSs)
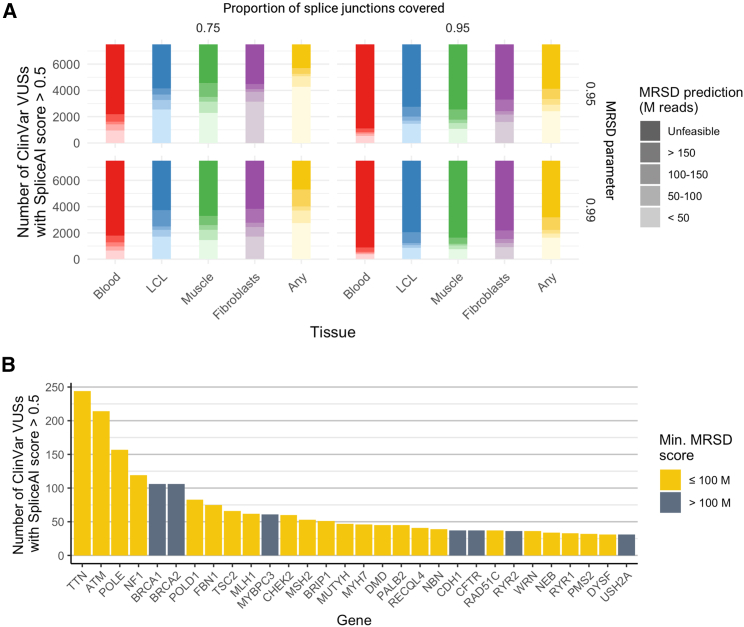
MRSD scores were derived for genes harboring VUSs present in ClinVar if the variants were predicted by SpliceAI to impact splicing (score ≥ 0.5; Jaganathan et al.22).
(A) Between 25.8% (1,940/7,507) and 67.8% (5,086/7,507) of variants predicted to impact splicing are expected to be adequately covered by 100 M uniquely mapping reads or fewer in at least one of the four tissues (whole blood, LCLs, skeletal muscle, and fibroblasts), dependent on model stringency. Variants were most likely to be found to be in low-MRSD genes (MRSD ≤ 100 M) in fibroblasts, irrespective of model parameters.
(B) Among the 30 genes with the greatest number of predicted splice-impacting VUSs, 23 were predicted to be adequately covered (using default parameters) with 100 M uniquely mapping reads or fewer in at least one of the four tissues. An 8-read junction support parameter was used throughout.