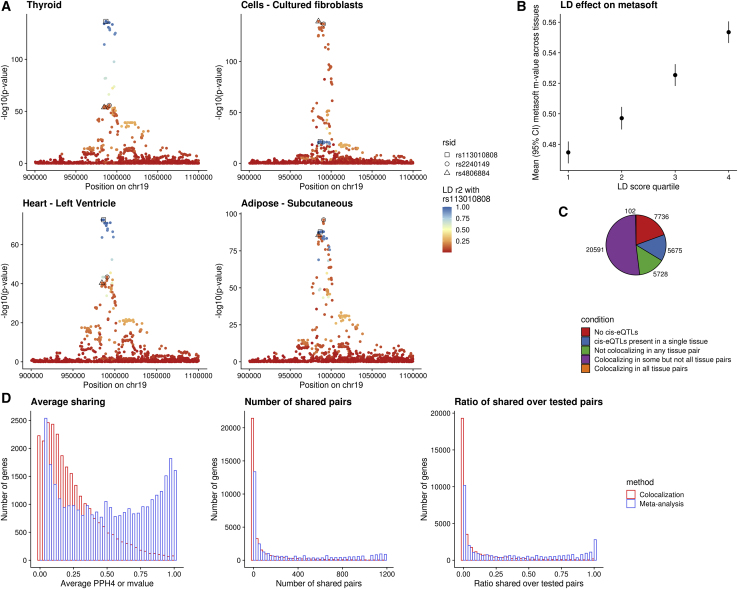
Figure 1.
Colocalization provides evidence of extensive eQTL tissue specificity
(A) Local Manhattan plots of cis-eQTLs for the WDR18 gene in four tissues of GTEx v8. The plots reveal allelic heterogeneity between tissues (while thyroid and heart share the same pattern of genetic regulation, whole blood and fibroblasts have different causal variants). The three lead variants across the four tissues are colored on the basis of their LD with variant rs113010808 (lead variant in both thyroid and heart). All three variants have a Metasoft m-value of 1 in all four tissues. LD r2 between rs2240149 and rs4806884 is 0.79, while r2 between rs2240149 and rs113010808 is 0.15.
(B) Metasoft m-values are positively correlated with LD. When the LD score quantile of the tested variant increases, the average m-value across tissues is higher.
(C) Pie-chart of all 38,518 genes in GTEx v8 that are expressed in at least one tissue based on the tissue specificity of their eQTLs estimated by COLOC.
(D) Histograms depicting patterns of sharing of genetic regulation between tissues based on COLOC and Metasoft. From left to right: histogram of mean PPH4 or m-value between tissue pairs for each gene; histogram of number of tissue pairs with shared cis-eQTLs for each gene; and histogram of the ratio of shared tissue pairs divided by the tissue pairs in which the gene is expressed. COLOC reveals more profound tissue specificity.