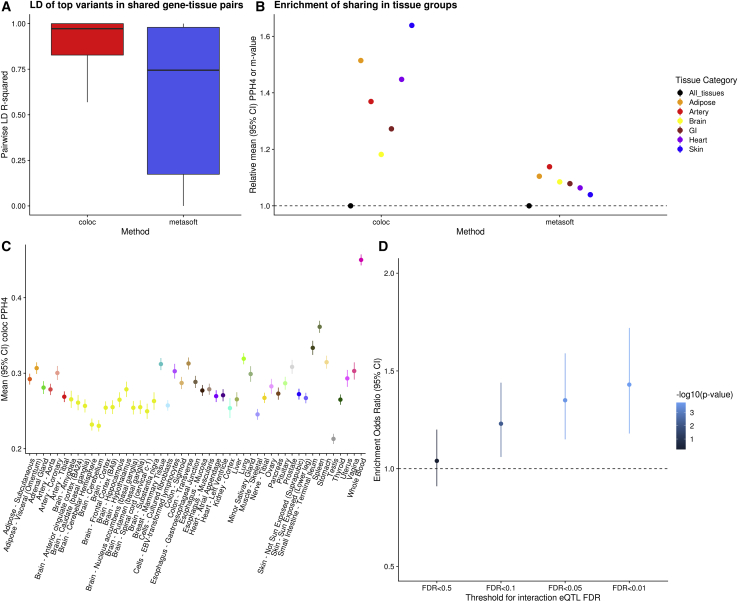
Figure 2.
Colocalization is superior to other methods that don’t account for LD
(A) Boxplots of the LD between the top variants per gene in tissue pairs that colocalize based on COLOC (in red) or Metasoft (in blue).
(B) Average sharing between groups of biologically similar tissues in GTEx v8 based on COLOC or Metasoft. Values are normalized by dividing with the mean across all gene-tissue pairs.
(C) Average eQTL sharing between eQTLGen whole blood and all GTEx v8 tissues for genes that have an eQTL in both datasets, using COLOC PPH4.
(D) Odds ratio of enrichment for non-colocalizing genes (PPH3 > 0.9) compared to colocalizing genes (PPH4 > 0.9) between eQTLGen whole blood and GTEx whole blood. The odds ratios are stratified by the false discovery rate of interaction with cell type composition, measured as the interaction between the lead eQTL variant for that gene in GTEx v8 and the relative proportion of neutrophils (the most common whole blood cell type) in the GTEx sample.