Figure 4.
Characteristics of genes based on the degree of eQTL tissue sharing
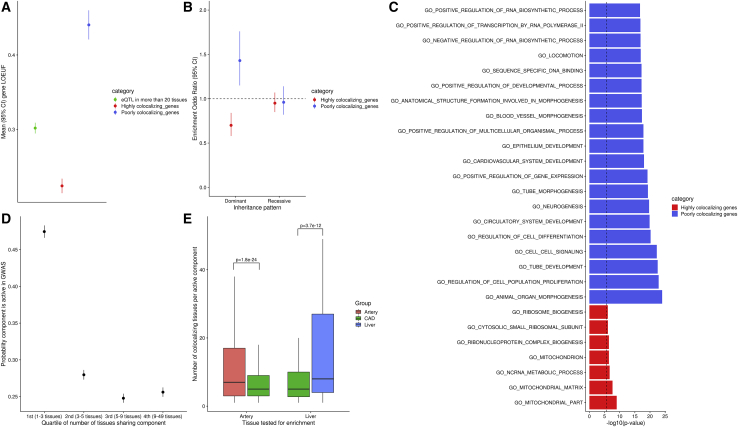
(A) Average loss of function observed/expected upper bound (LOEUF) between genes that are colocalizing in at least 20 tissues (highly colocalizing) and those that colocalize in less than five tissues (poorly colocalizing), comparing only genes that have an eQTL in at least 20 tissues. Colocalization was defined as sharing of the top causal component based on CAFEH.
(B) Enrichment in genes whose rare variation is associated with autosomal dominant or recessive human diseases based on OMIM (left) between highly and poorly colocalizing genes.
(C) Gene Ontology enrichment analysis of genes that are colocalizing in at least 20 tissues (highly colocalizing) and those that colocalize in less than five tissues (poorly colocalizing), comparing only genes that have an eQTL in at least 20 tissues. Colocalization was defined as sharing of the top causal component based on CAFEH.
(D) Mean (95% CI) of probability of a cis-eQTL component’s being active in GWAS (as defined by CAFEH-S) stratified by the quartile of the number of 49 GTEx tissues that share the given eQTL. The figure presents aggregate results of 19 diverse GWAS traits from the UK Biobank. For each GWAS, we jointly run CAFEH-S between the GWAS summary statistics and GTEx v8 eQTL summary statistics among genes for which the sentinel variant of a genome-wide significant disease locus is also a genome-wide eQTL in at least one tissue. We then evaluate all components that are active in at least one GTEx tissue. Overall, there is a strong inverse association between the posterior probability of a component being active in the GWAS and the number of tissues that colocalize with said component (linear mixed model p = 3.3 × 10−160).
(E) Boxplots of the number of tissues that share an active component identified by CAFEH, among components that are either active in a CAD GWAS and a GTEx tissue a priori believed to be relevant for CAD heritability (either artery on the left or liver on the right) among all genes within 1 Mb of genome-wide significant CAD locus, compared to components that are active only in a GTEx tissue (artery or liver) among all protein-coding genes.