Figure 5.
CAFEH identifies the target tissue in GWAS
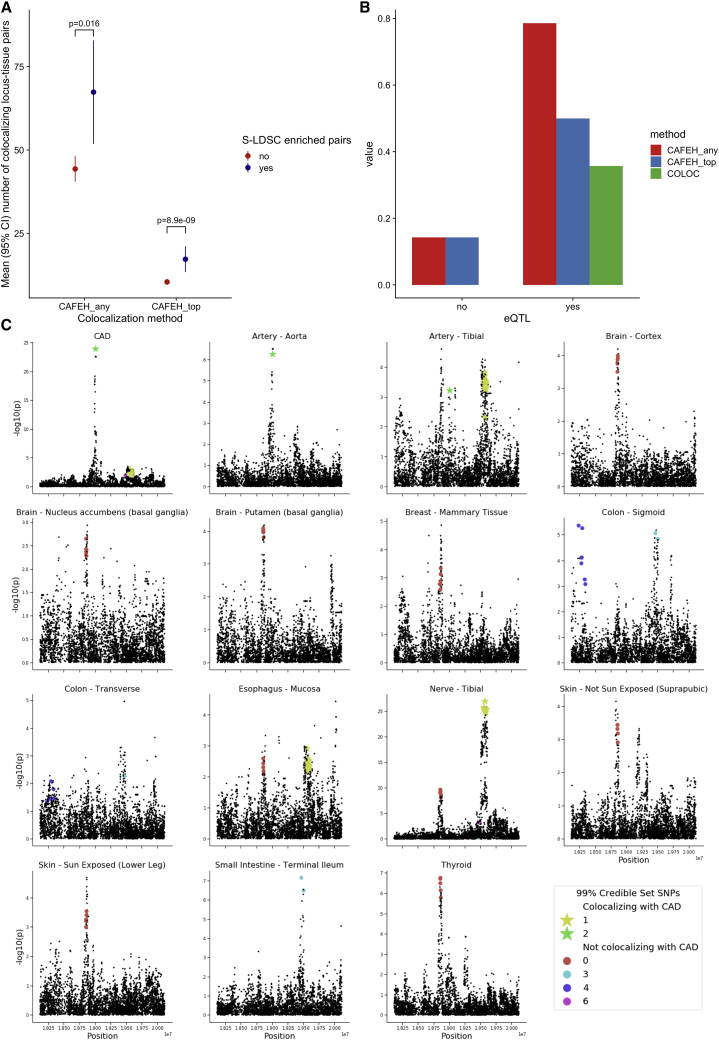
(A) Number of colocalizing locus tissue pairs in 19 diverse GWAS traits from the UK Biobank. Colocalization is defined on the basis of any component or top component in CAFEH and the counts are colored on the basis of whether the corresponding tissues are enriched for that trait by partitioned LD score regression.
(B) Case study of loci in a large CAD GWAS that have an established target tissue. The bars represent the proportion of loci colocalizing in the known target tissue with any of three different methods (CAFEH colocalization in any component, CAFEH colocalization in the top component, COLOC). The results are stratified on the basis of whether or not the lead variant in the locus is a genome-wide significant cis-eQTL for any gene in at least one tissue in GTEx v8. We see that CAFEH outperforms COLOC and can prioritize the target tissue in most cases when an eQTL signal is present.
(C) Example of CAFEH revealing allelic heterogeneity and identifying the target tissue in the TWIST1 CAD locus. CAFEH identifies a dominant component for the CAD GWAS in that locus with a 99% credible set that consists of a single variant (rs2107595). The same component is shared by artery tissue in GTEx v8. Recent CRISPR in human arterial smooth muscle cells confirmed its effects in influencing the expression of TWIST1 in that tissue.