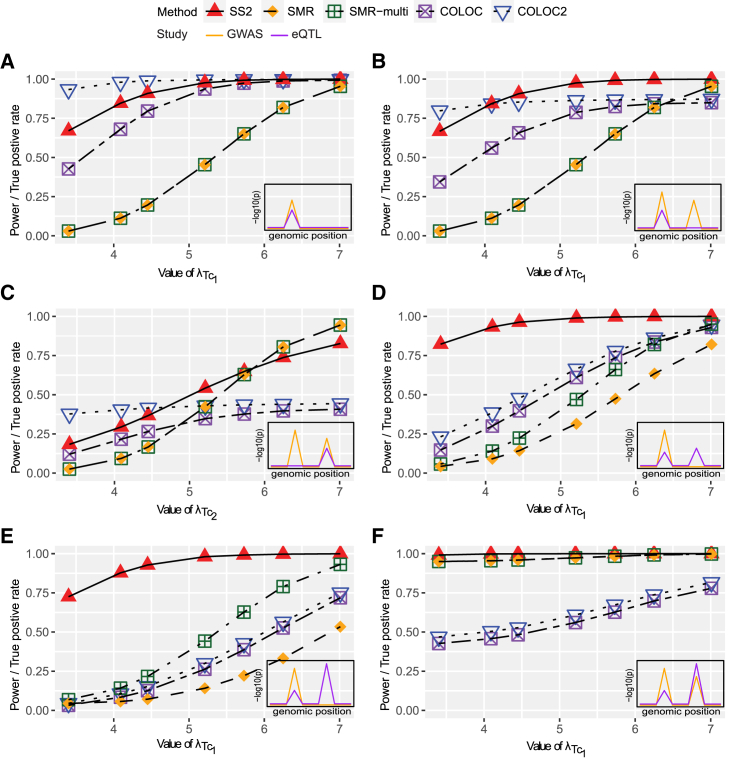
Figure 2.
Empirical power of SS2, SMR, and SMR-multi and true positive rates of COLOC and COLOC2 for testing a single gene for colocalization under six alternative scenarios: (A) one shared SNP for GWAS and eQTL study, (B) two independent GWAS SNPs, and one eQTL SNP is shared with the first GWAS SNP, (C) two independent GWAS SNPs, and one eQTL SNP is shared with the second GWAS SNP, (D) two independent eQTL SNPs, one GWAS SNP colocalizes with the eQTL SNP, given the non-overlapped eQTL association is low, (E) two independent eQTL SNPs, one GWAS SNP colocalizes with the eQTL SNP, given the non-overlapped eQTL association is strong, and (F) two independent GWAS SNPs, two independent eQTL SNPs and both SNPs are shared.
The LD pattern at the simulated region follows that at the MUC20/MUC4 locus at chromosome 3. For SS2, SMR,11 and SMR-multi,12 the nominal type 1 error rate is set at . For COLOC214 and COLOC,4 the false positive rates are controlled by applying the default 0.8 threshold (as recommended in Dobbyn et al.14) for the colocalization posterior probability. In total, 104 replications are simulated to obtain the empirical power and true positive rates. The x-axis demonstrates the parameter values for or which is set to be 3.4, 4.09, 4.45, 5.21, 5.73, or 7.01 such that 0.01, 0.05, 0.1, 0.3, 0.5, or 0.9 power is achieved to detect the eQTL association at the significance level of 10−8. The inserted figure in the right bottom of each plot provides the general visualization of GWAS (orange line) and eQTL (purple line) colocalization patterns in a region of interest.