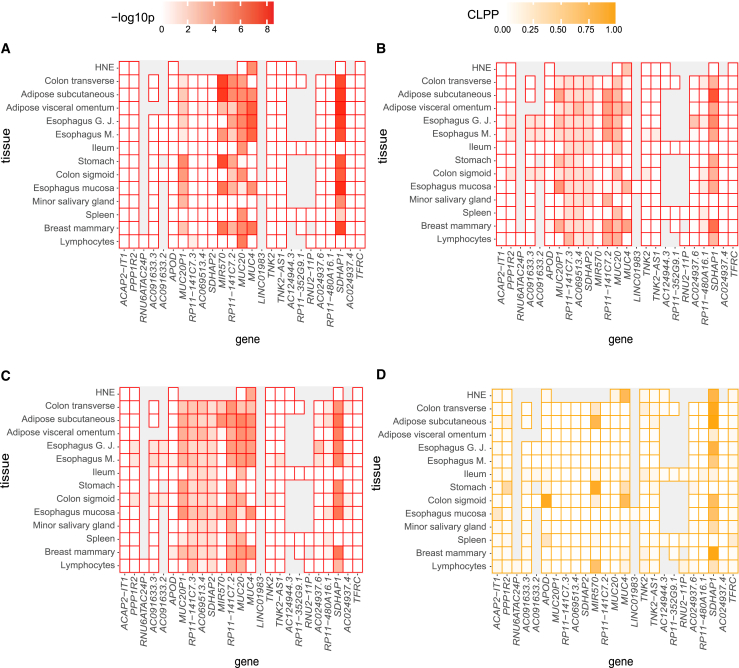
Figure 4.
Heatmaps of colocalization evidence across genes and tissues at chromosome 3
In each panel, each cell shows the colocalization evidence for the specified tissue and gene calculated from SNPs within 0.1 Mb of the GWAS peak variant. Genes on the x axis are annotated by GENCODE v.26 for hg38 GTEX V8 and are ordered by their chromosomal positions. For illustration purposes, only the closest annotated genes centered around the GWAS peak variant are shown, and all genes analyzed within 1 Mb around the GWAS peak variant is shown in Figure S2. Grey indicates insufficient expression levels attained for the gene in the tissue under study.
(A) The color intensity corresponds to the SS2 colocalization evidence as measured by –log10(SS2 p value), with red representing –log10(p) = 8.5 and white representing eQTL evidence for the corresponding gene and tissue does not pass the stage 1 test.
(B and C) The color intensity corresponds to the SMR and SMR-multi colocalization evidence as measured by –log10(SMR p value) and –log10(SMR-multi p value), respectively; with red representing –log10(p) = 8.5 and white representing eQTL evidence for the corresponding gene and tissue does not pass the eQTL p value threshold (5 × 10−8).
(D) The color intensity corresponds to the COLOC colocalization evidence as measured by colocalization posterior probability (CLPP) ranging from 0 to 1. The eQTL analyses used for all gene/tissue pairs are those conducted by GTEx27 v.8 release, except for the HNE eQTLs. eQTL analyses in HNE is conducted using FastQTL49 with RNA-sequencing of HNE from 94 CF-affected Canadians enrolled in the Canadian CF Gene Modifier study. Esophagus G.J represents Esophagus Gastresophageal Junction; Esophagus M. represents Esophagus Muscularis; Ileum represents Small Intestine Terminal Ileum; Lymphocytes represents cells EBV-transformed lymphocytes.