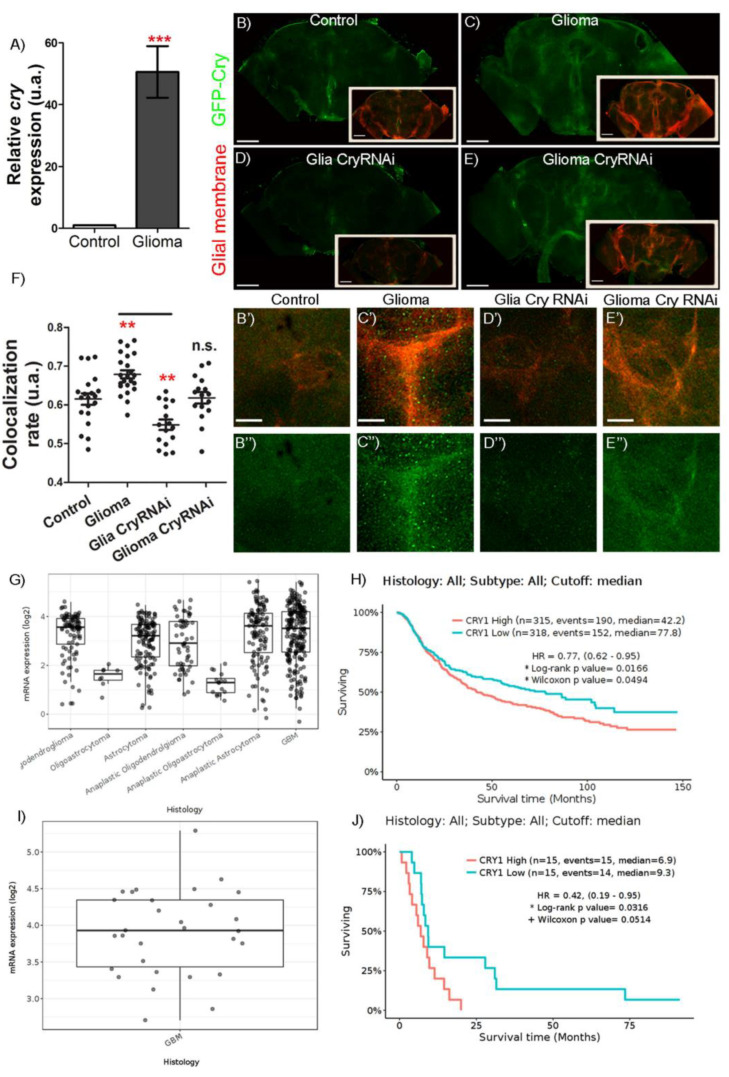
Figure 1.
Circadian gene cry is upregulated in both human GB samples and GB Drosophila model. (A) RT-qPCR analysis of complete brains of 7-day-old adult flies from repo-Gal4 > UAS-LacZ (Control) and repo-Gal4 > UAS-dEGFRλ, UAS-dp110 CAAX (Glioma) genotypes in LD conditions at ZT6 for the circadian gene cry (t-test) in n = 90. (B–E) Confocal microscopy images of brains of 7-day-old adult flies from (B) repo-Gal4 > UAS-LacZ (Control), (C) repo-Gal4 > UAS-dEGFRλ, UAS-dp110 CAAX (Glioma), (D) repo-Gal4 > UAS-dEGFRλ, UAS-dp110 CAAX, UAS-cryRNAi (Glioma CryRNAi) and (E) repo-Gal4 > UAS-cryRNAi (Glia CryRNAi) after using (B’–E″) magnifications of the brain lobe central region, the reporter GFP-Cry in green and the glial membrane are marked in red. (F) Co-localization between GFP-Cry signal and the glial membrane (mRFP). Statistical analysis in at least n = 16 (ANOVA, post-hoc Bonferroni). (G) Data on overexpression of cry1 in human primary gliomas and GB against normal tissue. (H) Graph showing a lower life expectancy in those patients with primary GB and cry1 overexpressed compared to patients with primary GB with low expression of cry1. (I) Data on cry1 expression in human secondary GB compared to normal tissue. (J) Graph showing a lower life expectancy in those patients with secondary GB and cry1 overexpressed compared to patients with secondary GB with low expression of cry1. Images obtained from gliovis.bioinfo.cnio.es based on the 2016 classification of brain tumors (scale bar, 100 µm in (B–E) and 20 µm in (B’–E’) (n.s. not significant, ** p-value < 0.01, *** p-value < 0.001).