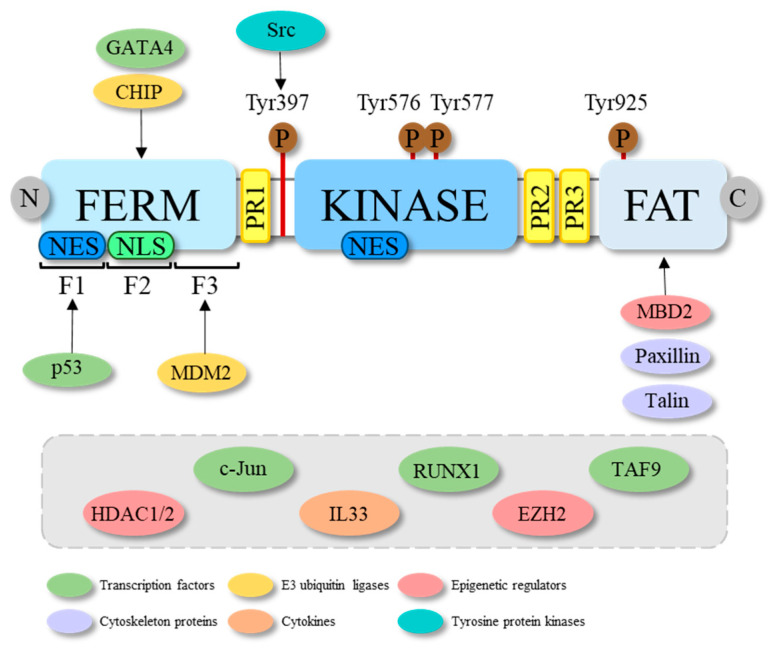
Figure 1.
Schematic representation of the molecular structure of FAK and some of its interactors. FAK structure consists of three main domains: the N-terminal FERM, the central kinase and the C-terminal FAT domain. The FERM domain includes three lobes, i.e., F1, F2 and F3, which are bound from transcription factors (among which are GATA4 and p53) and E3 ubiquitin ligases (among which are CHIP and MDM2). While it is known that p53 and MDM2, respectively, bind the F1 and F3 lobes of the FERM of FAK, for GATA4 and CHIP it is, only known that they bind the FERM domain, but unknown is to which of its three lobes. In the FERM domain a nuclear export signal (NES) in the lobe F1 and a nuclear localization signal (NLS) in the lobe F2, responsible for the nuclear export and localization of the protein, are reported. A NES is also found in the kinase domain. FAK structure further comprises three proline-rich (PR) regions that serve as binding sites for the Src homology (SH) 3 domains of several proteins. The main phosphorylation sites are shown with brown circles. In particular, the Tyr397 activation site, the Tyr576 and Tyr577 in the activation loop and the Tyr925 binding site for the SH2 domains are reported. The arrows indicate the FAK domain to which some of its interactors bind, while the grey rectangle shows the FAK interactors with still unknown binding sites.