Fig. 1 |. Three orthogonal screens identify CRL4AMBRA1 as a regulator of cyclin D1.
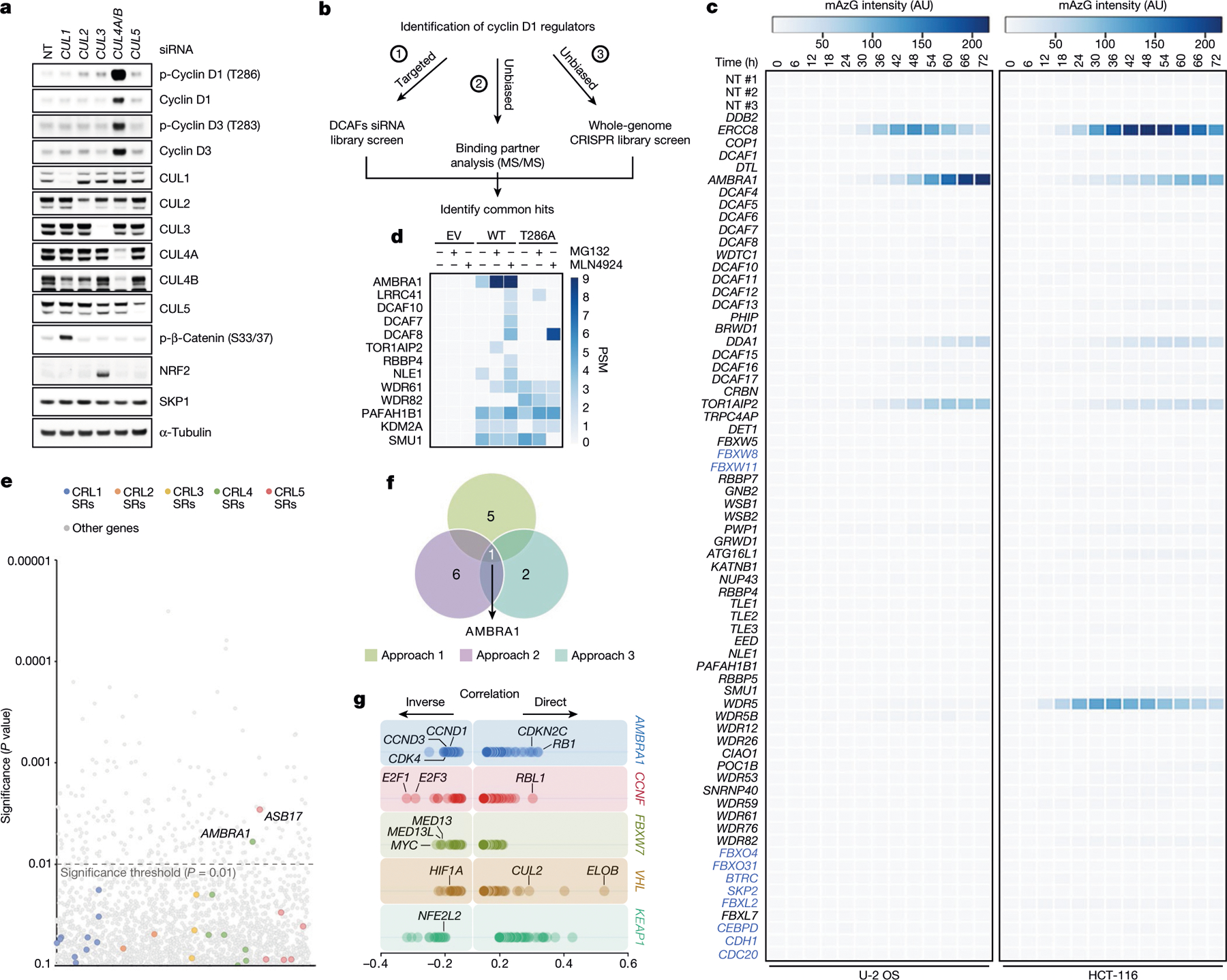
a, HCT-116 cells were transfected with a non-targeting (NT) siRNA or siRNAs against CUL1, CUL2, CUL3, CUL4A and CUL4B (CUL4A/B) or CUL5, and cell lysates were blotted with the indicated antibodies for proteins and phosphorylated (p-) proteins. This experiment was performed three times. b, Schematic representation of three orthogonal screens employed to identify regulators of cyclin D1. c, Heat map showing fluorescence intensity values in arbitrary units (AU), related to approach 1 in b. Genes encoding for the previously-reported regulators of D-type cyclins3 are reported in blue. This experiment was performed once in HCT-116 and once in U-2 OS cells. d, Heat map showing the number of peptide-spectrum matches (PSM) identified by LC-MS/MS matching substrate receptors from immunoprecipitates of empty vector (EV), wild-type (WT) cyclin D1, or cyclin D1(T286A), related to approach 2 in b. The full list of interactors is provided in Supplementary Table 1. e, Scatter plot of hits generated using approach 3 in b. This experiment was performed once, with three technical replicates. P values were calculated using the MAGeCK algorithm14. SR, substrate receptor. f, Venn diagram showing the number of hits obtained from the three screens in b. g, The top 100 CRISPR (Avana) Public 20Q1 pre-computed associations for AMBRA1 and selected substrate receptors that have a role in human cancer are depicted on the basis of their dependency profiles at genome scale (https://depmap.org/portal/; last accessed January 2020).
