Extended Data Fig. 2 |. Validation of candidate regulators of D-type cyclins.
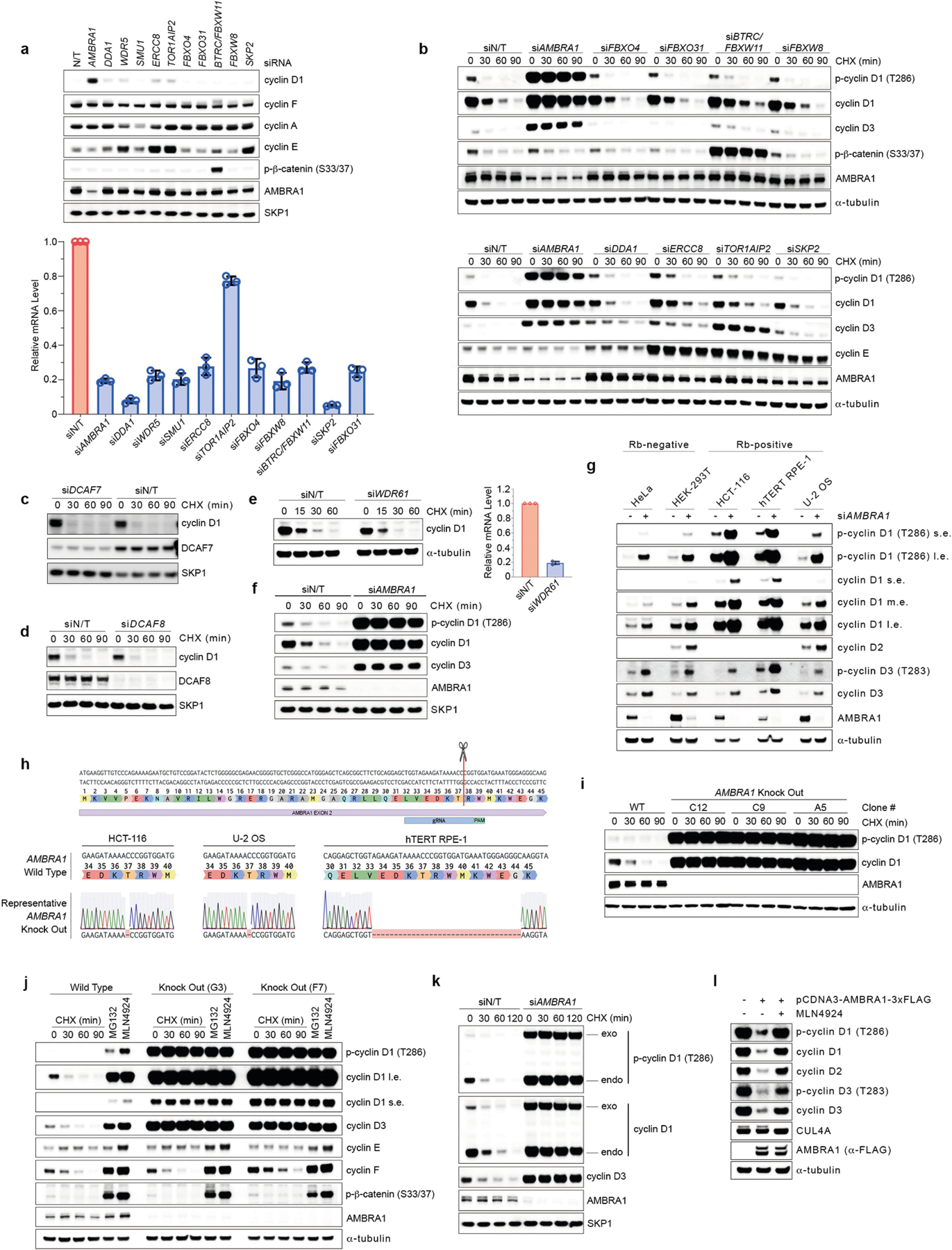
a, HCT-116 cells were transfected with a non-targeting siRNA or siRNAs against the indicated targets for two rounds before collection. Cell extracts were immunoblotted as indicated (upper panel). RNA extracts were also collected to measure the relative mRNA levels for each indicated target by quantitative PCR with reverse transcription (RT–qPCR). Each blue column represents the mRNA levels of the target indicated by the respective siRNA and normalized to its corresponding mRNA levels in cells transfected with a non-targeting siRNA (bottom). Data are mean mRNA levels ± s.d., n = 3 independent experiments. b, HCT-116 cells were transfected with a non-targeting siRNA or siRNAs against the indicated targets for two rounds followed by treatment with cycloheximide for the indicated times before collection. Cell extracts were immunoblotted as indicated. c, HCT-116 cells were transfected with a non-targeting siRNA or an siRNA against DCAF7 followed by treatment with cycloheximide for the indicated times before collection. Cell extracts were immunoblotted as indicated. d, U-2 OS cells were transfected with a non-targeting siRNA or an siRNA against DCAF8 followed by treatment with cycloheximide for the indicated times before collection. Cell extracts were immunoblotted as indicated. e, U-2 OS cells were transfected with a non-targeting siRNA or an siRNA against WDR61 followed by treatment with cycloheximide for the indicated times before collection. Cell extracts were immunoblotted as indicated (left). RNA extracts were also collected to measure the relative mRNA levels of WDR61 by RT–qPCR (right). Data are mean mRNA levels ± s.d., n = 3 independent experiments. f, hTERT RPE-1 cells were transfected with a non-targeting siRNA or an siRNA against AMBRA1 followed by treatment with cycloheximide for the indicated times before collection. Cell extracts were immunoblotted as indicated. g, HeLa, HEK 293T, HCT-116, hTERT RPE-1 and U-2 OS cells were transfected with a non-targeting siRNA or an siRNA against AMBRA1 before collection. Cell extracts were immunoblotted for the indicated proteins (l.e., long exposure; m.e., medium exposure; s.e., short exposure). h, Schematic representation of the AMBRA1 genomic locus and gRNA target location. Exon 2 refers to the human AMBRA1 gene in XM_005253009 (GRCh38. p13, NCBI gene: 55626). Representative AMBRA1 wild-type genomic DNA template and knockout mutant sequences identified by TOPO-TA cloning of PCR products from HCT-116, U-2 OS and hTERT-RPE1 cells are depicted. i, Three AMBRA1−/− and one AMBRA1+/+ hTERT RPE-1 clones were treated with cycloheximide for the indicated times, after which cell extracts were immunoblotted for the indicated proteins. j, Two AMBRA1−/− and one AMBRA1+/+ T98G clones were treated with either cycloheximide for the indicated times, MG132 or MLN4924 (both for 4 h) before collection. Cell extracts were then immunoblotted for the indicated proteins. k, HCT-116 cells were infected with a retrovirus expressing mAzG-CCND1 and transfected with a non-targeting siRNA or an siRNA against AMBRA1 for two rounds. Cells were then treated with cycloheximide for the indicated times, after which cell extracts were immunoblotted for the indicated proteins (endo, endogenous; exo, exogenous). l, AMBRA1−/− U-2 OS cells were transiently transfected with a vector expressing a C-terminal 3×Flag-tagged AMBRA1 or an empty vector for 96 h. Where indicated, cells were treated with MLN4924 for 4 h before lysis. Cell extracts were immunoblotted as indicated. Unless otherwise noted, experiments were performed at least three independent times.
