Extended Data Fig. 3 |. Depletion of AMBRA1 using the auxin-inducible degron system.
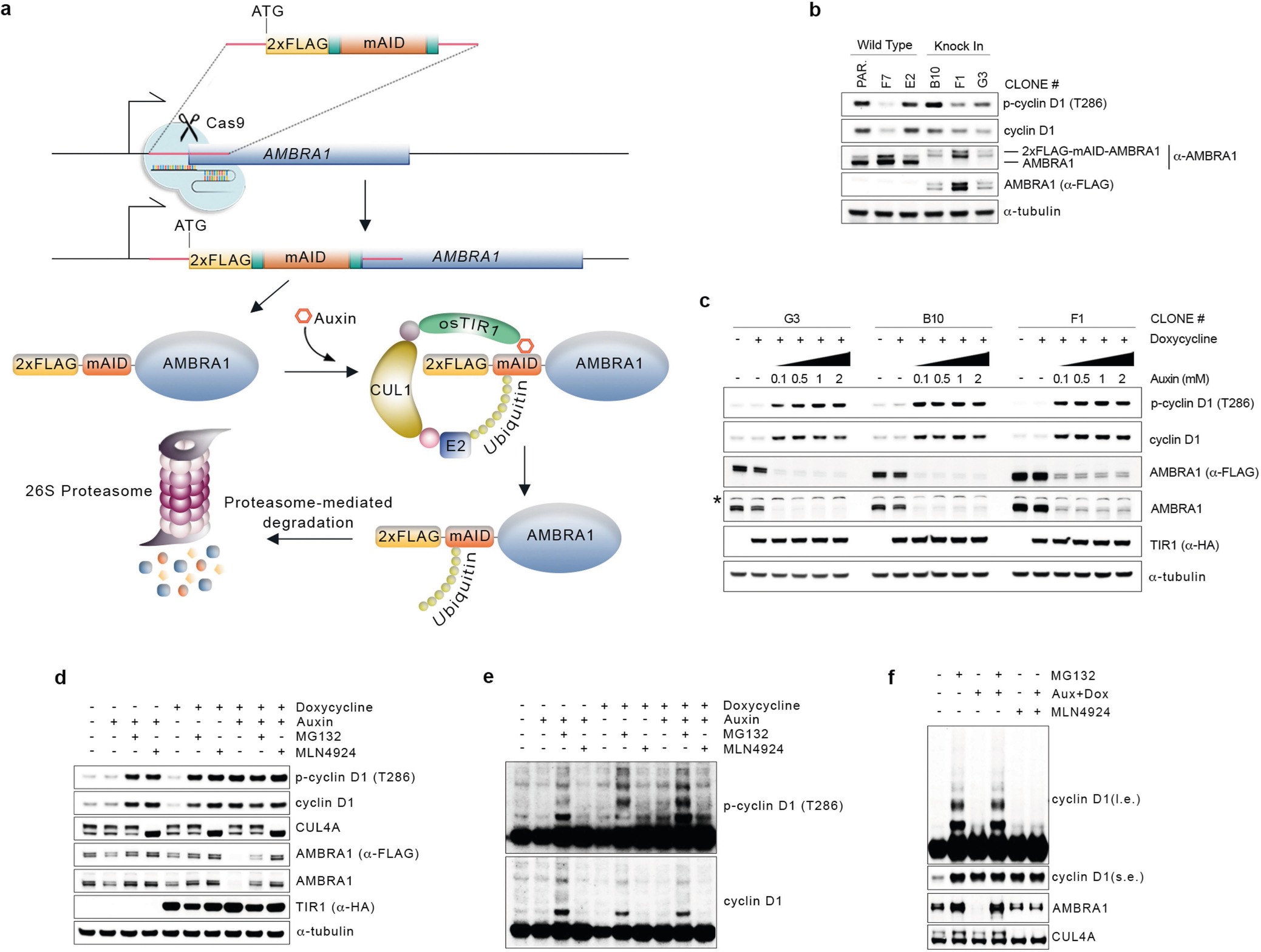
a, Schematic representation of the CRISPR–Cas9-based strategy to insert an N-terminal, 2×Flag-tagged minimal AID (mAID) in the endogenous locus of AMBRA1 (5′-end of exon 2) in HCT-116 cells. Cells were infected with a doxycycline-inducible lentivirus expressing Oryza sativa TIR1 (osTIR1). b, Three knock-in clones were obtained as in a and compared to wild-type clones for proper endogenous tagging of AMBRA1 by immunoblotting. Note the higher molecular weight of 2×Flag-mAID-tagged AMBRA1. c, Three knock-in clones were treated with doxycycline with or without increasing doses of auxin for 12 h. Cell extracts were immunoblotted as indicated. The asterisk indicates a non-specific band. d, One clone of 2×Flag-mAID-AMBRA1 HCT-116 cells (clone #F1) was treated with either DMSO or doxycycline for 12 h, and/or auxin, MG132 and MLN4924 (alone or in combination) for 4 h as indicated. Cell extracts were immunoblotted as indicated. e, Longer exposure from the cyclin D1 and phospho-cyclin D1 (T286) immunoblots described in d. f, Whole-cell extracts from 2×Flag-mAID-AMBRA1 HCT-116 cells pre-treated with either DMSO or doxycycline for 12 h, and exposed to combinations of auxin, MG132 and/or MLN4924 as indicated for 4 h, related to Fig. 2d. Cell extracts were immunoblotted as indicated. Unless otherwise noted, experiments were performed at least three independent times.
