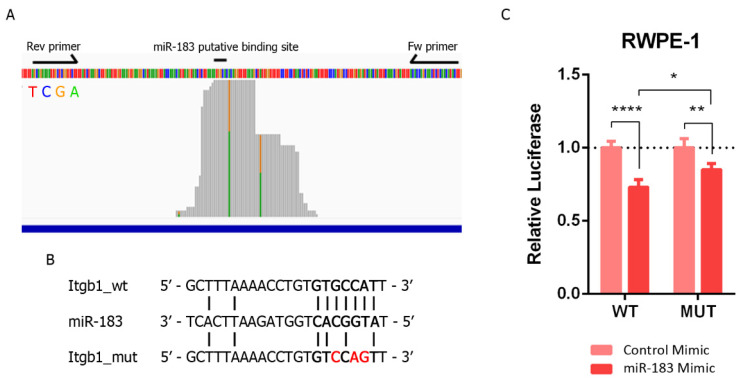
Figure 4.
Validation of the direct miR-183 and ITGB1 mRNA interaction in PrCa. (A) Mapping of prostate cell line reads along the putative binding site of miR-183 on ITGB1’s 3′UTR extracted from AGO-PAR-CLIP RNA-seq data [31]. The base identity is represented by four colors, as indicated. Perfect base matches and mismatches (due to base substitutions caused by the photoactivatable ribonucleoside crosslinking) are represented as gray and colored bars, respectively. Forward and reverse primers used to clone the miR-183 binding site of the ITGB1 3′UTR in pmiRGLO are indicated by arrows. The image is a modified screenshot of the IGV Software [42]. (B) Base pairing between miR-183 and the conserved miR-183 putative binding site on ITGB1 wild type and mutated 3′UTR cloned in pmirGLO for reporter gene assays. (C) Reporter gene assay using pmiRGLO, bearing the region of the 3′UTR of ITGB1 with the miR-183 wild type or mutated site, co-transfected with miR-183 mimic or control RNA in the RWPE-1 cell line. Luciferase activity was measured 48 h after transfection. The mean ± SD of four independent experiments is shown. A two-way ANOVA was applied to test the statistical significance of the differences between conditions. p-value * <0.05, ** <0.01, **** <0.0001.