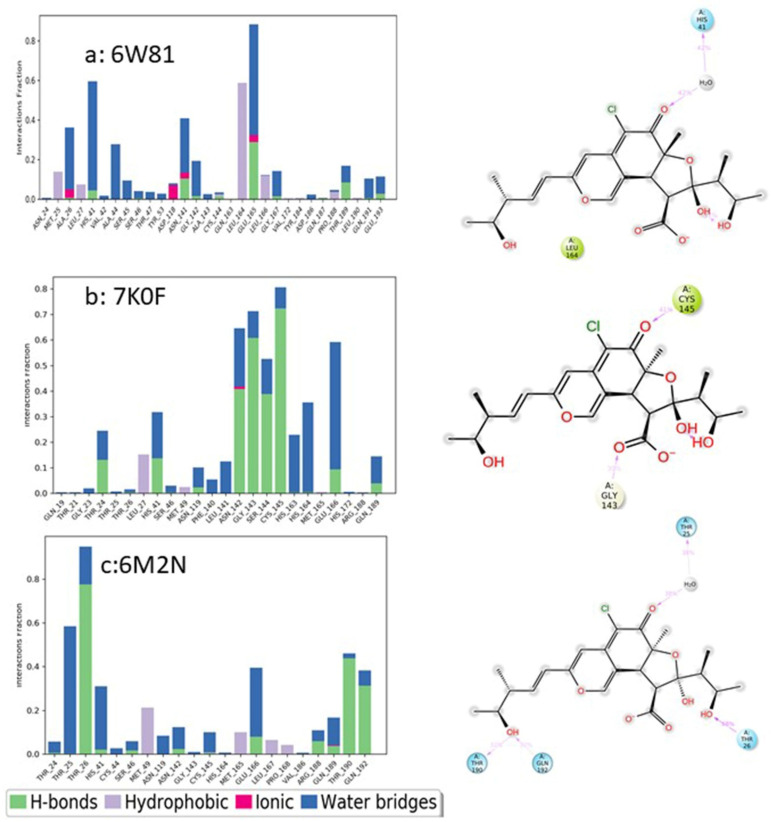
Figure 14.
Analysis of molecular interactions and types of contacts with 3CL pro-hydrolase during the MD simulations. Detailed schematic interactions of chaetovirdin D atoms with binding site residues of hydrolase crystal structures (a) 6W81, (b) 7K0F, and (c) 6M2N. Interactions happening during more than 30% of the simulation is shown. Normalized stacked bar charts show the viral protease binding site residues interacting with chaetovirdin D via hydrogen bonds, hydrophobic and ionic interactions, and water bridges.