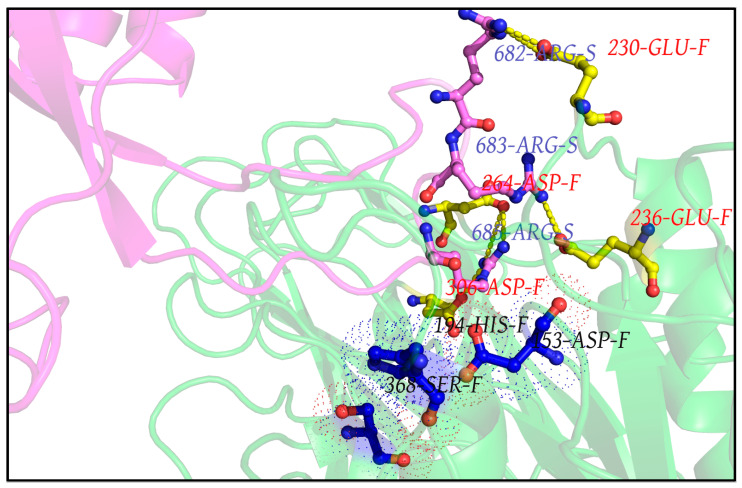
Figure 4.
The high persistence interfacial salt-bridges formed between FLCSSpike and the triad-proximal Furin anionic residues (in RR1CoV-2). The figure highlights the highest persistence Spike–Furin interfacial salt-bridges (thick yellow dashed lines) for the three arginines in the 681PRRAR685 (FLCSSpike) along its 300 ns MD simulation trajectory (produced from RR1CoV-2). Only a close up view of the interface is portrayed to highlight the key (highly persistent) salt-bridges (pers > 0.85) sustaining the interface. Only the Furin proximal part (FLCSSpike with short flanking regions at either end) of the Spike (chain S) is shown in magenta cartoon (with R682, R683, R685 highlighted in ‘balls and sticks’), while the Furin chain (chain F) is drawn in green cartoon with its counter-ionic residues involved in the (aforementioned) key Spike–Furin interfacial salt-bridges highlighted in ‘balls and sticks’. Residues comprising the catalytic triad are presented in ‘balls and sticks’ colored in navy blue and highlighted by dot surface points. Figures were built in PyMol. Residues are differentially labeled with font colors blue (681PRRAR685 arginines), red (Furin anionic residues) and black (catalytic triad).