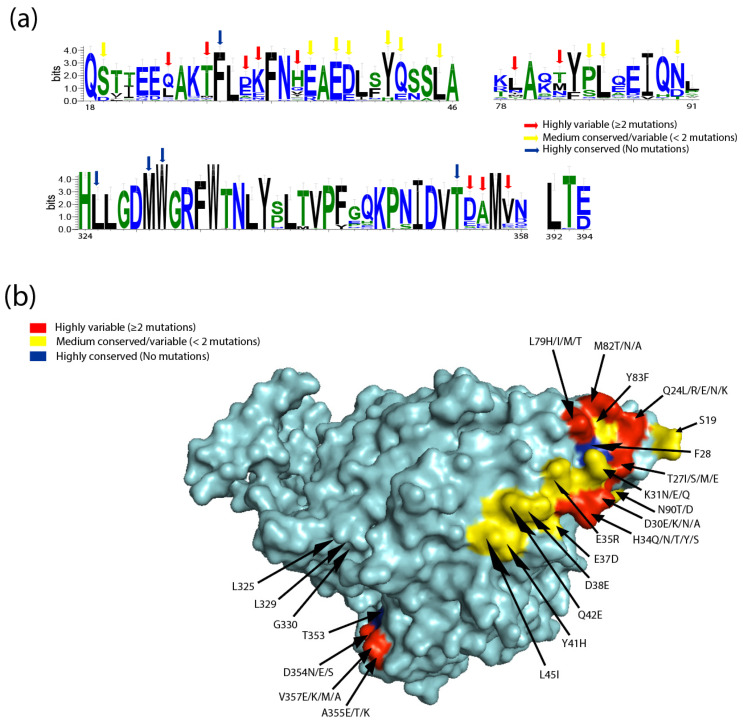
Figure 3.
The binding sites for SARS-CoV-2 on ACE2 show a high degree of variation. (a) WebLogo graphs illustrating the amino acid divergence between mammalian and the ACE2 sequences of different species. The vertical height of the amino acid (aa 18–46, 78–91, 324–358, and 392–394) represents its predominance at each location in the polypeptide (aa 18–46, 78–91, 324–358, and 392–394). WebLogo (University of California, Berkeley, USA) [57] plots summarizing the amino acid divergence within the mammalian and ACE2 sequences of the different species included in this study. (b) Conservation of mammalian ACE2 amino acid residues, estimated from site-specific evolutionary rates, mapped onto the surface of the ACE2 ectodomain, and coloured: red (highly variable (≥2 mutations)), yellow (medium conserved/variable (<2 mutations)), and blue (highly conserved (No mutations)). Inset depicts the SARS-CoV-2 binding region of ACE2, with residues that contact the SARS-CoV-2 RBD highlighted.